Gene
KWMTBOMO13075 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000302
Annotation
PREDICTED:_constitutive_coactivator_of_PPAR-gamma-like_protein_1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.092 Nuclear Reliability : 1.552
Sequence
CDS
ATGGGCATACAAGACTTACAAACATTTTTAGAAAGTGAAGGTGTAGGTGTTGATCTCTTTAAAATTGCGAGAAACCATTCTGGAGGTTTGCGCCTGGTTCTCGACGCCGAAGGTTGTTTGGATCGTCTTTACGGTGGATACTTCTCAGATTGGGCAAGTGGTGGACAATGGGCCCGATGTGTACAGTTCTTAACAACACTAGCACAAGCTTTGGCTGAATTTCAAGTATCACTTTGTGTTTGTTTTAATGGATCTCATCCTACGCCTCCTGCATTACCCCAACATTGGATCCAGACTCAAACCATATATAAGCAAAGAGTGAATTCAGTTTTGCACCATATTACCACTAAAGGTACTCCACCACCTAAAGTATGGTGGGTACCACCTACTGGACTGCATTCATGTTTACGAACCGCACTTAGGTACCTCAATATCCCAATTATGGTGTCTTCTGATGATCACTGTCAGGAAGTTGTTGGTTTATGTCGTGAAAAATCATATGCTGGCCTCGTTGGATGCTCTGGAGAGTATTTAGTGTTTCAACCACCTAGATACTTTAGTTCATCTTTGTTGAAACTAACATATAGATCTTCACTAGAAACTAAAGAATATATGGTCCATGATTTGCCACAAGTACTTAATGTACCTCAAGATCGTCTTTGCCTCATGGCTGCATTACTTGGAGGAGTCATTCTTTCAGAACAAAGCTTGACAGATTTCTATAAACGCCTTGGATTAGTGCCCAAAAAGCAAGTTCCTCCAGAGATACAAATAAAAACAATTTGTCAGTATGTAAGTGAATTACCAACTTCTGATGTTGATGCAGTCTGCATAGATATATTTGGAGATCTGAATGACCTACGCGCTGCAAAACTTAAACAAGCCGTACAGTATTATCTAAACGGTACAAAGGATGGTTTCTTAAAATATAAAACAGCTACAGGTCGTCGTTCAAAAAATAAAAAGAATCTCCAAAGACAAGAATTCAACATGCAGCCTGTTACTCTGGATCTAGATACATCAAAATTAGCGACTGAATCGTCAGAACATGAGCAAAAAGGCTTGGAAGATTACAAATTGGCTACAGCAAATGCATCAATGAGTGCTGACTTTAGTATGGAGAAACAAACTAATGACTTACATCATGGATTCGCATCTCTATCACTTGAAGCTGGTGTTATGTCATCTACTAAGCAACAAAAACAGAATATTATCAAAAAAAAGGCTGAAAAACCACAATCTGCAATTAAAGAGGGGCGACAAGCTACAAAACAAGTCCAAAGTGATGACAACTGCCCTCCAGCACCGGCAGAAGTATTGCGGACCTGTGCGGAACGTCACACCAGGGGCCTCATGCATCCTCAACTTCTATCTATCCTTACACATCGTCAAGTGACTTTACCTGTGCTTATGGAGGATGAGAATCATAGAGAAATTCCTTCAATTCATCATTTCTTTAGACCAATTCGACAGAATGTATATGCTATTCTGTACAATATGCATCATCATCGCTTTATGGCCCACAAAATTAAAGATGATAGATCGAGTGGCTCTAATGATCGTCCATGCACAGTTTTGATAGCAGAATGGATATACACCCGCACAAATCCTGGCAAGAATCCAGAGATTGTTCCAGGTGTTTTACTGGATTGGCCTGTGCCAACTGTACAGAGGCTCTGGTTTGGAACTGCTCCGGATGATAAATGCCGTAGAATGAGAGCATATCTTTCCTGTATGCGCTCAGATACTCCTCTGATGTTGAATACTACTTATGTGCCCCAGCACTTGCTCATACTGGCAACTGTATTGAGATTTATAATGACATCTCCAATTCAAATACTACGAAGGCAAGAGCTGGATGCATTTTTAGCTACTGCATTTTCGACAGATCTTATGAATGCACTACATTTGCAAGAAATGAATATCAATGCTATCAGTTCCCGGGGCGTACAATTAGGCGCGCTGGTGATGGCGGGCGTGGAGGCGGCGCTCGTGGCTAATGACGCTTGCGGTGCTCCTGTACCATGGCTCGTAGCTTCACCCTGGCTCTACTTCGATGGCAAGCTGCTGCAGCGAAACCTCCATCGCGCAGCACACTGCAAACATCTTCCTCAGCTTTGCGATAACAACCTCGACATCGTCGTCAAGGTTGAGCGCATGCGAAAGGCGATCTTGGAAGGTCTGGAGGTGGAGTTCGCGATGGCGCCGCTGCCGCTCGTGGCGGGCGTGCTAGGGAGCGACGGGGGCACGTGGGGCGCGAGGGGGCGGGGCCGCGGCGGCCGGCTTGAGATCGCGGGCGTTGTCGTCGGACAGTGGGGCGGCGGCAGCTATCCCGCACGCGCTCCGCCCAGATACCAACAGATGAGCGCGCACACTGTGGGCTACGTTGGATACACGCCTCCGACTCGGAACTCGTACGGAGGTCGCGGGTGGCGCGGAGCACGCGGGGGTCGCGGCCGGGGACACGCGCGCCCCTCGCGCCGCCCCCGCCGGGACCCGAACGAGCCCGCCAAACCAGCCACTATTGCCGTTAATGGGAAAACCGTTCGCCCGGACGAAGTTCACCACCAGTCAGGTGAATCAAGTCCTCAAGATGAAAATACTGCTCAAGAAGGTGAAGCAGAAGGAACACCCACTCCTAAACCTAAAACTGGCAAGAATCTGAAGAAAAATGCAGGAAAGGGGAAAAAAAGGATCAATAAACCGGAAGGCTTGCAAGGAGCATCAGAGGCTAATGGACCGCTTGATAAAAATAAGGTGGCAGAGCAAGGCCACAGTTCCGACAAGGAACACATAGGGCAAGGTGATGCACCAGTCTCAAATTAA
Protein
MGIQDLQTFLESEGVGVDLFKIARNHSGGLRLVLDAEGCLDRLYGGYFSDWASGGQWARCVQFLTTLAQALAEFQVSLCVCFNGSHPTPPALPQHWIQTQTIYKQRVNSVLHHITTKGTPPPKVWWVPPTGLHSCLRTALRYLNIPIMVSSDDHCQEVVGLCREKSYAGLVGCSGEYLVFQPPRYFSSSLLKLTYRSSLETKEYMVHDLPQVLNVPQDRLCLMAALLGGVILSEQSLTDFYKRLGLVPKKQVPPEIQIKTICQYVSELPTSDVDAVCIDIFGDLNDLRAAKLKQAVQYYLNGTKDGFLKYKTATGRRSKNKKNLQRQEFNMQPVTLDLDTSKLATESSEHEQKGLEDYKLATANASMSADFSMEKQTNDLHHGFASLSLEAGVMSSTKQQKQNIIKKKAEKPQSAIKEGRQATKQVQSDDNCPPAPAEVLRTCAERHTRGLMHPQLLSILTHRQVTLPVLMEDENHREIPSIHHFFRPIRQNVYAILYNMHHHRFMAHKIKDDRSSGSNDRPCTVLIAEWIYTRTNPGKNPEIVPGVLLDWPVPTVQRLWFGTAPDDKCRRMRAYLSCMRSDTPLMLNTTYVPQHLLILATVLRFIMTSPIQILRRQELDAFLATAFSTDLMNALHLQEMNINAISSRGVQLGALVMAGVEAALVANDACGAPVPWLVASPWLYFDGKLLQRNLHRAAHCKHLPQLCDNNLDIVVKVERMRKAILEGLEVEFAMAPLPLVAGVLGSDGGTWGARGRGRGGRLEIAGVVVGQWGGGSYPARAPPRYQQMSAHTVGYVGYTPPTRNSYGGRGWRGARGGRGRGHARPSRRPRRDPNEPAKPATIAVNGKTVRPDEVHHQSGESSPQDENTAQEGEAEGTPTPKPKTGKNLKKNAGKGKKRINKPEGLQGASEANGPLDKNKVAEQGHSSDKEHIGQGDAPVSN
Summary
Uniprot
H9ISS5
A0A2H1V248
A0A2W1BQU6
A0A194Q367
A0A3S2LYT0
S4PZQ7
+ More
A0A2A4JEZ2 A0A194R1P0 A0A212F494 A0A3L8D7X1 A0A154PRP2 A0A026WWN9 A0A195FBP7 E2C9D6 A0A0L7QSJ0 A0A151WRG7 A0A151JB26 F4WWE0 A0A0C9QKY4 A0A151IJQ6 A0A195BM27 A0A158NKU7 A0A2A3EEN6 A0A087ZSS8 V9IF34 A0A0M9A7R7 E9IFE3 A0A0J7L437 E2AMV3 N6TFM2 A0A1B6MJD2 A0A1B6IUQ0 A0A1Y1LHL4 A0A1B6DEG4 A0A1B6BZX8 A0A232ET60 D6X382 A0A067RC80 E0W1G9 A0A310SPD5 T1INJ6 A0A131XSI3 A0A2R5LND0 A0A147BT08 A0A224XIS9 A0A293LFH4 A0A1Z5KUY9 A0A069DZP8 A0A023EZ11 A0A1B6CTA4 A0A164XMC5 A0A0P5C165 A0A0P5Y6G7 A0A0P5USN4 A0A0P5CDK8 K7J5D3 A0A0P5HR18 A0A2S2R7U2 A0A0P6ESD0 K1PD57 A0A2H8THJ7 V4B249 J9K5B8 A0A1L8D8K5 A0A1L8D867 A0A1L8D8E7 A0A336KB12 A0A1J1I404 R7TXK4 A0A1D2MSJ9 A0A1W2W5H8
A0A2A4JEZ2 A0A194R1P0 A0A212F494 A0A3L8D7X1 A0A154PRP2 A0A026WWN9 A0A195FBP7 E2C9D6 A0A0L7QSJ0 A0A151WRG7 A0A151JB26 F4WWE0 A0A0C9QKY4 A0A151IJQ6 A0A195BM27 A0A158NKU7 A0A2A3EEN6 A0A087ZSS8 V9IF34 A0A0M9A7R7 E9IFE3 A0A0J7L437 E2AMV3 N6TFM2 A0A1B6MJD2 A0A1B6IUQ0 A0A1Y1LHL4 A0A1B6DEG4 A0A1B6BZX8 A0A232ET60 D6X382 A0A067RC80 E0W1G9 A0A310SPD5 T1INJ6 A0A131XSI3 A0A2R5LND0 A0A147BT08 A0A224XIS9 A0A293LFH4 A0A1Z5KUY9 A0A069DZP8 A0A023EZ11 A0A1B6CTA4 A0A164XMC5 A0A0P5C165 A0A0P5Y6G7 A0A0P5USN4 A0A0P5CDK8 K7J5D3 A0A0P5HR18 A0A2S2R7U2 A0A0P6ESD0 K1PD57 A0A2H8THJ7 V4B249 J9K5B8 A0A1L8D8K5 A0A1L8D867 A0A1L8D8E7 A0A336KB12 A0A1J1I404 R7TXK4 A0A1D2MSJ9 A0A1W2W5H8
Pubmed
EMBL
BABH01013180
BABH01013181
BABH01013182
BABH01013183
ODYU01000339
SOQ34917.1
+ More
KZ150009 PZC75120.1 KQ459586 KPI97850.1 RSAL01000111 RVE47107.1 GAIX01001118 JAA91442.1 NWSH01001742 PCG70268.1 KQ460883 KPJ11434.1 AGBW02010414 OWR48555.1 QOIP01000011 RLU16580.1 KQ435090 KZC14572.1 KK107105 EZA59549.1 KQ981693 KYN37806.1 GL453806 EFN75481.1 KQ414756 KOC61607.1 KQ982805 KYQ50499.1 KQ979182 KYN22307.1 GL888406 EGI61482.1 GBYB01004199 JAG73966.1 KQ977294 KYN03880.1 KQ976439 KYM86779.1 ADTU01018872 ADTU01018873 ADTU01018874 KZ288266 PBC30197.1 JR039984 JR039985 AEY58934.1 AEY58935.1 KQ435718 KOX78970.1 GL762838 EFZ20692.1 LBMM01000718 KMQ97652.1 GL440936 EFN65227.1 APGK01029502 APGK01029503 KB740735 KB631805 ENN79169.1 ERL86280.1 GEBQ01003953 JAT36024.1 GECU01017051 JAS90655.1 GEZM01055107 GEZM01055106 GEZM01055105 GEZM01055104 JAV73142.1 GEDC01031325 GEDC01028651 GEDC01022805 GEDC01016758 GEDC01013214 GEDC01005936 JAS05973.1 JAS08647.1 JAS14493.1 JAS20540.1 JAS24084.1 JAS31362.1 GEDC01030729 GEDC01022302 GEDC01007130 GEDC01006626 JAS06569.1 JAS14996.1 JAS30168.1 JAS30672.1 NNAY01002323 OXU21550.1 KQ971372 EFA09788.1 KK852801 KDR16324.1 DS235870 EEB19475.1 KQ760505 OAD60064.1 JH431178 GEFM01006766 JAP69030.1 GGLE01006873 MBY10999.1 GEGO01001889 JAR93515.1 GFTR01008487 JAW07939.1 GFWV01002524 MAA27254.1 GFJQ02008048 JAV98921.1 GBGD01000265 JAC88624.1 GBBI01004269 JAC14443.1 GEDC01020687 JAS16611.1 LRGB01000966 KZS14381.1 GDIP01181824 JAJ41578.1 GDIP01062386 JAM41329.1 GDIP01108931 JAL94783.1 GDIP01188074 JAJ35328.1 AAZX01001890 AAZX01009687 AAZX01019915 GDIQ01224449 JAK27276.1 GGMS01016557 MBY85760.1 GDIQ01071839 JAN22898.1 JH818358 EKC21787.1 GFXV01001724 MBW13529.1 KB203946 ESO82329.1 ABLF02034459 ABLF02034466 GFDF01011430 JAV02654.1 GFDF01011431 JAV02653.1 GFDF01011429 JAV02655.1 UFQS01000156 UFQT01000156 SSX00615.1 SSX20995.1 CVRI01000040 CRK95071.1 AMQN01002390 KB309044 ELT95705.1 LJIJ01000612 ODM95868.1
KZ150009 PZC75120.1 KQ459586 KPI97850.1 RSAL01000111 RVE47107.1 GAIX01001118 JAA91442.1 NWSH01001742 PCG70268.1 KQ460883 KPJ11434.1 AGBW02010414 OWR48555.1 QOIP01000011 RLU16580.1 KQ435090 KZC14572.1 KK107105 EZA59549.1 KQ981693 KYN37806.1 GL453806 EFN75481.1 KQ414756 KOC61607.1 KQ982805 KYQ50499.1 KQ979182 KYN22307.1 GL888406 EGI61482.1 GBYB01004199 JAG73966.1 KQ977294 KYN03880.1 KQ976439 KYM86779.1 ADTU01018872 ADTU01018873 ADTU01018874 KZ288266 PBC30197.1 JR039984 JR039985 AEY58934.1 AEY58935.1 KQ435718 KOX78970.1 GL762838 EFZ20692.1 LBMM01000718 KMQ97652.1 GL440936 EFN65227.1 APGK01029502 APGK01029503 KB740735 KB631805 ENN79169.1 ERL86280.1 GEBQ01003953 JAT36024.1 GECU01017051 JAS90655.1 GEZM01055107 GEZM01055106 GEZM01055105 GEZM01055104 JAV73142.1 GEDC01031325 GEDC01028651 GEDC01022805 GEDC01016758 GEDC01013214 GEDC01005936 JAS05973.1 JAS08647.1 JAS14493.1 JAS20540.1 JAS24084.1 JAS31362.1 GEDC01030729 GEDC01022302 GEDC01007130 GEDC01006626 JAS06569.1 JAS14996.1 JAS30168.1 JAS30672.1 NNAY01002323 OXU21550.1 KQ971372 EFA09788.1 KK852801 KDR16324.1 DS235870 EEB19475.1 KQ760505 OAD60064.1 JH431178 GEFM01006766 JAP69030.1 GGLE01006873 MBY10999.1 GEGO01001889 JAR93515.1 GFTR01008487 JAW07939.1 GFWV01002524 MAA27254.1 GFJQ02008048 JAV98921.1 GBGD01000265 JAC88624.1 GBBI01004269 JAC14443.1 GEDC01020687 JAS16611.1 LRGB01000966 KZS14381.1 GDIP01181824 JAJ41578.1 GDIP01062386 JAM41329.1 GDIP01108931 JAL94783.1 GDIP01188074 JAJ35328.1 AAZX01001890 AAZX01009687 AAZX01019915 GDIQ01224449 JAK27276.1 GGMS01016557 MBY85760.1 GDIQ01071839 JAN22898.1 JH818358 EKC21787.1 GFXV01001724 MBW13529.1 KB203946 ESO82329.1 ABLF02034459 ABLF02034466 GFDF01011430 JAV02654.1 GFDF01011431 JAV02653.1 GFDF01011429 JAV02655.1 UFQS01000156 UFQT01000156 SSX00615.1 SSX20995.1 CVRI01000040 CRK95071.1 AMQN01002390 KB309044 ELT95705.1 LJIJ01000612 ODM95868.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000218220
UP000053240
UP000007151
+ More
UP000279307 UP000076502 UP000053097 UP000078541 UP000008237 UP000053825 UP000075809 UP000078492 UP000007755 UP000078542 UP000078540 UP000005205 UP000242457 UP000005203 UP000053105 UP000036403 UP000000311 UP000019118 UP000030742 UP000215335 UP000007266 UP000027135 UP000009046 UP000076858 UP000002358 UP000005408 UP000030746 UP000007819 UP000183832 UP000014760 UP000094527
UP000279307 UP000076502 UP000053097 UP000078541 UP000008237 UP000053825 UP000075809 UP000078492 UP000007755 UP000078542 UP000078540 UP000005205 UP000242457 UP000005203 UP000053105 UP000036403 UP000000311 UP000019118 UP000030742 UP000215335 UP000007266 UP000027135 UP000009046 UP000076858 UP000002358 UP000005408 UP000030746 UP000007819 UP000183832 UP000014760 UP000094527
PRIDE
SUPFAM
SSF88723
SSF88723
ProteinModelPortal
H9ISS5
A0A2H1V248
A0A2W1BQU6
A0A194Q367
A0A3S2LYT0
S4PZQ7
+ More
A0A2A4JEZ2 A0A194R1P0 A0A212F494 A0A3L8D7X1 A0A154PRP2 A0A026WWN9 A0A195FBP7 E2C9D6 A0A0L7QSJ0 A0A151WRG7 A0A151JB26 F4WWE0 A0A0C9QKY4 A0A151IJQ6 A0A195BM27 A0A158NKU7 A0A2A3EEN6 A0A087ZSS8 V9IF34 A0A0M9A7R7 E9IFE3 A0A0J7L437 E2AMV3 N6TFM2 A0A1B6MJD2 A0A1B6IUQ0 A0A1Y1LHL4 A0A1B6DEG4 A0A1B6BZX8 A0A232ET60 D6X382 A0A067RC80 E0W1G9 A0A310SPD5 T1INJ6 A0A131XSI3 A0A2R5LND0 A0A147BT08 A0A224XIS9 A0A293LFH4 A0A1Z5KUY9 A0A069DZP8 A0A023EZ11 A0A1B6CTA4 A0A164XMC5 A0A0P5C165 A0A0P5Y6G7 A0A0P5USN4 A0A0P5CDK8 K7J5D3 A0A0P5HR18 A0A2S2R7U2 A0A0P6ESD0 K1PD57 A0A2H8THJ7 V4B249 J9K5B8 A0A1L8D8K5 A0A1L8D867 A0A1L8D8E7 A0A336KB12 A0A1J1I404 R7TXK4 A0A1D2MSJ9 A0A1W2W5H8
A0A2A4JEZ2 A0A194R1P0 A0A212F494 A0A3L8D7X1 A0A154PRP2 A0A026WWN9 A0A195FBP7 E2C9D6 A0A0L7QSJ0 A0A151WRG7 A0A151JB26 F4WWE0 A0A0C9QKY4 A0A151IJQ6 A0A195BM27 A0A158NKU7 A0A2A3EEN6 A0A087ZSS8 V9IF34 A0A0M9A7R7 E9IFE3 A0A0J7L437 E2AMV3 N6TFM2 A0A1B6MJD2 A0A1B6IUQ0 A0A1Y1LHL4 A0A1B6DEG4 A0A1B6BZX8 A0A232ET60 D6X382 A0A067RC80 E0W1G9 A0A310SPD5 T1INJ6 A0A131XSI3 A0A2R5LND0 A0A147BT08 A0A224XIS9 A0A293LFH4 A0A1Z5KUY9 A0A069DZP8 A0A023EZ11 A0A1B6CTA4 A0A164XMC5 A0A0P5C165 A0A0P5Y6G7 A0A0P5USN4 A0A0P5CDK8 K7J5D3 A0A0P5HR18 A0A2S2R7U2 A0A0P6ESD0 K1PD57 A0A2H8THJ7 V4B249 J9K5B8 A0A1L8D8K5 A0A1L8D867 A0A1L8D8E7 A0A336KB12 A0A1J1I404 R7TXK4 A0A1D2MSJ9 A0A1W2W5H8
Ontologies
GO
PANTHER
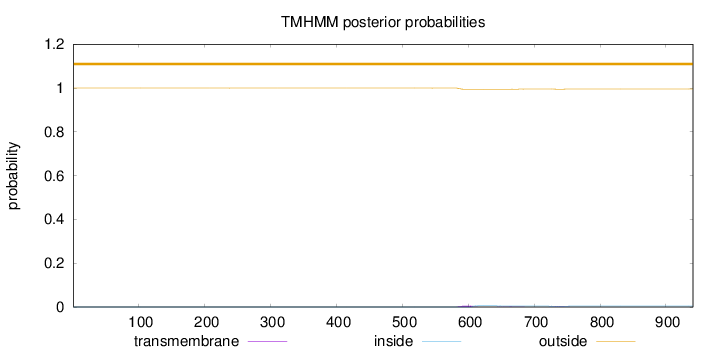
Topology
Length:
941
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19437
Exp number, first 60 AAs:
4e-05
Total prob of N-in:
0.00022
outside
1 - 941
Population Genetic Test Statistics
Pi
419.231118
Theta
198.091415
Tajima's D
3.050692
CLR
0.3437
CSRT
0.981350932453377
Interpretation
Uncertain
