Pre Gene Modal
BGIBMGA000397
Annotation
RGS-GAIP_interacting_protein_GIPC_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.185 Nuclear Reliability : 1.582
Sequence
CDS
ATGTCGTTATTTAAAAAGAAAGCGCCTAAATACACGCCGCCACCGGTACCACCCACACCAGTAGAAGATACTGAATCACAGTCTAATGGAGTCGATAATAGCAACGGTACGCAGAAGTCACAACTGGTCTTCCACTGTCAGCAAGCTCACGGAAGTCCATTGGGCCTCATCTCTGGTTTCTCTAACGTCAAAGAATTATATGAGAAGATAGCAGAATGTTACGAATTTTCACCCGAAGATATTCTTTTCTGTACTTTAAATACTCACAAAGTGGATATGAAAAAACTTCTCGGAGGGCAAATAGGATTAGAAGACTTTATTTTCGCGCATAGAAAAGGTCGACCAAAGGAAATAGAAATAGTTAAAACAGAAGATGCTCTAGGATTAACGATTACAGATAACGGAGCAGGTTATGCATTTATAAAAAGGATTAAAGAAGGCTCCATTGTTTCAAGAATACCACATATTGAGGTCGGAGATCATATTGAGAAATTGAATGGTGAGAATATGGTTGGGAAAAGACATTATGAGGTGGCGAAAATTCTGAAAGACATACCGAAGGGAACCACTTTTATAATGCGATTGGTGGAACCATTGAAGAGTGGATTCGGTAGCATCGGTCCTAAAACCGGTAGTGGGAGGTCAGGAAAAAAACAAAACTTTGGCTCAGGAAAAGAAACTCTACGTTTTAAAGCGAATGGCCAAGCAACAATTGAAAATGAGCACGATGACAAATCAAAGGCTGGAATTGAAAAAATCAACGGTCTACTGGAATCATTTATGGGTATCAATGATAGTGAATTGGCATCGCAAATGTGGGATCTTGCCGAAGGAAAACAAAATTCTATGGAACTCGCAGAGGCTATCGATAACAGCGATCTTCAAGAATTTGGTTTTACAGATGAATTTATCATTGAACTATGGGGAGTTATAACTGATGCTAGATCTGGAAGACTCAGTTAG
Protein
MSLFKKKAPKYTPPPVPPTPVEDTESQSNGVDNSNGTQKSQLVFHCQQAHGSPLGLISGFSNVKELYEKIAECYEFSPEDILFCTLNTHKVDMKKLLGGQIGLEDFIFAHRKGRPKEIEIVKTEDALGLTITDNGAGYAFIKRIKEGSIVSRIPHIEVGDHIEKLNGENMVGKRHYEVAKILKDIPKGTTFIMRLVEPLKSGFGSIGPKTGSGRSGKKQNFGSGKETLRFKANGQATIENEHDDKSKAGIEKINGLLESFMGINDSELASQMWDLAEGKQNSMELAEAIDNSDLQEFGFTDEFIIELWGVITDARSGRLS
Summary
Uniprot
Q2F696
A0A1E1WHH9
A0A3S2L716
A0A212F496
A0A2W1BTQ8
A0A194R1P4
+ More
S4PGM5 H9IT18 A0A0L7KUY4 A0A0N7Z9V8 A0A194PYL8 A0A0K8TKE9 A0A182Q4W4 A0A182XZF0 A0A2M4AHP5 A0A2M4AGQ8 A0A182UV85 Q7QJV3 D6X0C9 A0A034VY92 A0A023EQA6 A0A1B0CMS5 A0A1Q3G5A1 T1E9Z3 A0A1Q3G5C2 A0A336M7P8 A0A0A1XCS0 A0A2M4CRK6 A0A2M4BSI6 A0A1L8DMA5 A0A2M4CRN3 W8BJF0 A0A2M3Z208 V5GRE7 A0A182IJL8 E9HF44 A0A084VF71 A0A0P5YJ64 A0A1S3DUX0 A0A2H1V422 A0A1Y1KAR2 A0A1L8DM09 A0A0J9R825 Q7JX82 B4HRK6 A0A0P6JS04 B4NMR5 B4P2P1 B3N8T6 A0A1J1J3W2 A0A1W4VYY5 A0A1J1J6L5 B4KNW1 B3MGT9 B4GGM9 Q28ZX6 A0A3Q0II38 A0A3Q0INK3 A0A3B0J282 T1PBJ3 A0A026W172 E2C237 A0A1B6EAC6 A0A1A9V8H2 A0A1B0FQG0 A0A1A9Z5J2 A0A1B0BV49 A0A1A9WP09 A0A1B6LBY5 B4J8C4 A0A1A9XB70 A0A158NUJ8 A0A0M5J337 B4LJN2 A0A1I8PYU9 T1I230 A0A224XSQ1 J3JVI6 A0A1B6FM06 A0A023F3Y3 A0A2H8TLD2 C4WSV7 A0A087U047 A0A146L901 A0A146KSW3 A0A0N0U608 A0A151X6L2 J9JKJ2 A0A2L2Y1Q7 A0A2S2Q945 A0A195C9C2 E2A382 A0A0P4VNI6 A0A195FIY5 C1C1V8 A0A087ZSQ2 A0A151J2S6 A0A0K2V9I6 A0A0K2V8F8 A0A2A3EEJ6
S4PGM5 H9IT18 A0A0L7KUY4 A0A0N7Z9V8 A0A194PYL8 A0A0K8TKE9 A0A182Q4W4 A0A182XZF0 A0A2M4AHP5 A0A2M4AGQ8 A0A182UV85 Q7QJV3 D6X0C9 A0A034VY92 A0A023EQA6 A0A1B0CMS5 A0A1Q3G5A1 T1E9Z3 A0A1Q3G5C2 A0A336M7P8 A0A0A1XCS0 A0A2M4CRK6 A0A2M4BSI6 A0A1L8DMA5 A0A2M4CRN3 W8BJF0 A0A2M3Z208 V5GRE7 A0A182IJL8 E9HF44 A0A084VF71 A0A0P5YJ64 A0A1S3DUX0 A0A2H1V422 A0A1Y1KAR2 A0A1L8DM09 A0A0J9R825 Q7JX82 B4HRK6 A0A0P6JS04 B4NMR5 B4P2P1 B3N8T6 A0A1J1J3W2 A0A1W4VYY5 A0A1J1J6L5 B4KNW1 B3MGT9 B4GGM9 Q28ZX6 A0A3Q0II38 A0A3Q0INK3 A0A3B0J282 T1PBJ3 A0A026W172 E2C237 A0A1B6EAC6 A0A1A9V8H2 A0A1B0FQG0 A0A1A9Z5J2 A0A1B0BV49 A0A1A9WP09 A0A1B6LBY5 B4J8C4 A0A1A9XB70 A0A158NUJ8 A0A0M5J337 B4LJN2 A0A1I8PYU9 T1I230 A0A224XSQ1 J3JVI6 A0A1B6FM06 A0A023F3Y3 A0A2H8TLD2 C4WSV7 A0A087U047 A0A146L901 A0A146KSW3 A0A0N0U608 A0A151X6L2 J9JKJ2 A0A2L2Y1Q7 A0A2S2Q945 A0A195C9C2 E2A382 A0A0P4VNI6 A0A195FIY5 C1C1V8 A0A087ZSQ2 A0A151J2S6 A0A0K2V9I6 A0A0K2V8F8 A0A2A3EEJ6
Pubmed
22118469
28756777
26354079
23622113
19121390
26227816
+ More
26369729 25244985 12364791 14747013 17210077 18362917 19820115 25348373 24945155 25830018 24495485 21292972 24438588 28004739 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 26999592 17550304 18057021 15632085 23185243 25315136 24508170 30249741 20798317 21347285 22516182 25474469 26823975 26561354 27129103
26369729 25244985 12364791 14747013 17210077 18362917 19820115 25348373 24945155 25830018 24495485 21292972 24438588 28004739 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17994087 26999592 17550304 18057021 15632085 23185243 25315136 24508170 30249741 20798317 21347285 22516182 25474469 26823975 26561354 27129103
EMBL
DQ311176
ABD36121.1
GDQN01004733
GDQN01004107
JAT86321.1
JAT86947.1
+ More
RSAL01000111 RVE47111.1 AGBW02010414 OWR48551.1 KZ150009 PZC75123.1 KQ460883 KPJ11439.1 GAIX01006145 JAA86415.1 BABH01013185 JTDY01005372 KOB67067.1 GDRN01109108 JAI57128.1 KQ459586 KPI97844.1 GDAI01002844 JAI14759.1 AXCN02000591 GGFK01006994 MBW40315.1 GGFK01006648 MBW39969.1 AAAB01008807 EAA04121.4 EDO64513.1 KQ971372 EFA09593.1 GAKP01012120 GAKP01012117 GAKP01012116 JAC46835.1 GAPW01002348 JAC11250.1 AJWK01019146 GFDL01000070 JAV34975.1 GAMD01000867 JAB00724.1 GFDL01000076 JAV34969.1 UFQT01000518 SSX24899.1 GBXI01012033 GBXI01006379 GBXI01006039 JAD02259.1 JAD07913.1 JAD08253.1 GGFL01003786 MBW67964.1 GGFJ01006831 MBW55972.1 GFDF01006587 JAV07497.1 GGFL01003785 MBW67963.1 GAMC01009435 GAMC01009434 GAMC01009433 GAMC01009432 JAB97123.1 GGFM01001816 MBW22567.1 GALX01004284 JAB64182.1 GL732633 EFX69627.1 ATLV01012364 KE524785 KFB36615.1 GDIP01057001 LRGB01003375 JAM46714.1 KZS03021.1 ODYU01000396 SOQ35112.1 GEZM01087696 JAV58529.1 GFDF01006586 JAV07498.1 CM002911 KMY92216.1 AE013599 AY060451 AY095011 AAF59122.1 AAF59123.1 AAF59124.1 AAG22301.1 AAL25490.1 AAM11339.1 AHN55985.1 CH480816 EDW46888.1 GDUN01000806 JAN95113.1 CH964282 EDW85654.1 CM000157 EDW89302.1 KRJ98359.1 KRJ98360.1 CH954177 EDV59563.1 CVRI01000070 CRL07125.1 CRL07126.1 CH933808 EDW09006.1 KRG04418.1 KRG04419.1 CH902619 EDV37857.1 KPU77059.1 KPU77060.1 CH479183 EDW35649.1 CM000071 EAL25486.2 KRT02266.1 KRT02267.1 KRT02268.1 KRT02269.1 OUUW01000001 SPP75227.1 KA646132 AFP60761.1 KK107496 QOIP01000004 EZA49812.1 RLU24089.1 GL452067 EFN77988.1 GEDC01002419 JAS34879.1 CCAG010008865 JXJN01021141 GEBQ01018772 JAT21205.1 CH916367 EDW02283.1 ADTU01026585 CP012524 ALC42468.1 CH940648 EDW61600.1 ACPB03000298 GFTR01004956 JAW11470.1 BT127254 AEE62216.1 GECZ01018533 JAS51236.1 GBBI01002552 JAC16160.1 GFXV01002677 MBW14482.1 AK340352 BAH70977.1 KK117538 KFM70736.1 GDHC01013775 JAQ04854.1 GDHC01019211 GDHC01015116 JAP99417.1 JAQ03513.1 KQ435760 KOX75574.1 KQ982481 KYQ55997.1 ABLF02024065 ABLF02024069 ABLF02024070 ABLF02043444 IAAA01005079 LAA01230.1 GGMS01005050 MBY74253.1 KQ978143 KYM96713.1 GL436311 EFN72110.1 GDKW01003271 JAI53324.1 KQ981522 KYN40625.1 BT080837 ACO15261.1 KQ980368 KYN16330.1 HACA01029461 CDW46822.1 HACA01029462 CDW46823.1 KZ288266 PBC30157.1
RSAL01000111 RVE47111.1 AGBW02010414 OWR48551.1 KZ150009 PZC75123.1 KQ460883 KPJ11439.1 GAIX01006145 JAA86415.1 BABH01013185 JTDY01005372 KOB67067.1 GDRN01109108 JAI57128.1 KQ459586 KPI97844.1 GDAI01002844 JAI14759.1 AXCN02000591 GGFK01006994 MBW40315.1 GGFK01006648 MBW39969.1 AAAB01008807 EAA04121.4 EDO64513.1 KQ971372 EFA09593.1 GAKP01012120 GAKP01012117 GAKP01012116 JAC46835.1 GAPW01002348 JAC11250.1 AJWK01019146 GFDL01000070 JAV34975.1 GAMD01000867 JAB00724.1 GFDL01000076 JAV34969.1 UFQT01000518 SSX24899.1 GBXI01012033 GBXI01006379 GBXI01006039 JAD02259.1 JAD07913.1 JAD08253.1 GGFL01003786 MBW67964.1 GGFJ01006831 MBW55972.1 GFDF01006587 JAV07497.1 GGFL01003785 MBW67963.1 GAMC01009435 GAMC01009434 GAMC01009433 GAMC01009432 JAB97123.1 GGFM01001816 MBW22567.1 GALX01004284 JAB64182.1 GL732633 EFX69627.1 ATLV01012364 KE524785 KFB36615.1 GDIP01057001 LRGB01003375 JAM46714.1 KZS03021.1 ODYU01000396 SOQ35112.1 GEZM01087696 JAV58529.1 GFDF01006586 JAV07498.1 CM002911 KMY92216.1 AE013599 AY060451 AY095011 AAF59122.1 AAF59123.1 AAF59124.1 AAG22301.1 AAL25490.1 AAM11339.1 AHN55985.1 CH480816 EDW46888.1 GDUN01000806 JAN95113.1 CH964282 EDW85654.1 CM000157 EDW89302.1 KRJ98359.1 KRJ98360.1 CH954177 EDV59563.1 CVRI01000070 CRL07125.1 CRL07126.1 CH933808 EDW09006.1 KRG04418.1 KRG04419.1 CH902619 EDV37857.1 KPU77059.1 KPU77060.1 CH479183 EDW35649.1 CM000071 EAL25486.2 KRT02266.1 KRT02267.1 KRT02268.1 KRT02269.1 OUUW01000001 SPP75227.1 KA646132 AFP60761.1 KK107496 QOIP01000004 EZA49812.1 RLU24089.1 GL452067 EFN77988.1 GEDC01002419 JAS34879.1 CCAG010008865 JXJN01021141 GEBQ01018772 JAT21205.1 CH916367 EDW02283.1 ADTU01026585 CP012524 ALC42468.1 CH940648 EDW61600.1 ACPB03000298 GFTR01004956 JAW11470.1 BT127254 AEE62216.1 GECZ01018533 JAS51236.1 GBBI01002552 JAC16160.1 GFXV01002677 MBW14482.1 AK340352 BAH70977.1 KK117538 KFM70736.1 GDHC01013775 JAQ04854.1 GDHC01019211 GDHC01015116 JAP99417.1 JAQ03513.1 KQ435760 KOX75574.1 KQ982481 KYQ55997.1 ABLF02024065 ABLF02024069 ABLF02024070 ABLF02043444 IAAA01005079 LAA01230.1 GGMS01005050 MBY74253.1 KQ978143 KYM96713.1 GL436311 EFN72110.1 GDKW01003271 JAI53324.1 KQ981522 KYN40625.1 BT080837 ACO15261.1 KQ980368 KYN16330.1 HACA01029461 CDW46822.1 HACA01029462 CDW46823.1 KZ288266 PBC30157.1
Proteomes
UP000283053
UP000007151
UP000053240
UP000005204
UP000037510
UP000053268
+ More
UP000075886 UP000076408 UP000075903 UP000007062 UP000007266 UP000092461 UP000075880 UP000000305 UP000030765 UP000076858 UP000079169 UP000000803 UP000001292 UP000007798 UP000002282 UP000008711 UP000183832 UP000192221 UP000009192 UP000007801 UP000008744 UP000001819 UP000268350 UP000095301 UP000053097 UP000279307 UP000008237 UP000078200 UP000092444 UP000092445 UP000092460 UP000091820 UP000001070 UP000092443 UP000005205 UP000092553 UP000008792 UP000095300 UP000015103 UP000054359 UP000053105 UP000075809 UP000007819 UP000078542 UP000000311 UP000078541 UP000005203 UP000078492 UP000242457
UP000075886 UP000076408 UP000075903 UP000007062 UP000007266 UP000092461 UP000075880 UP000000305 UP000030765 UP000076858 UP000079169 UP000000803 UP000001292 UP000007798 UP000002282 UP000008711 UP000183832 UP000192221 UP000009192 UP000007801 UP000008744 UP000001819 UP000268350 UP000095301 UP000053097 UP000279307 UP000008237 UP000078200 UP000092444 UP000092445 UP000092460 UP000091820 UP000001070 UP000092443 UP000005205 UP000092553 UP000008792 UP000095300 UP000015103 UP000054359 UP000053105 UP000075809 UP000007819 UP000078542 UP000000311 UP000078541 UP000005203 UP000078492 UP000242457
Interpro
ProteinModelPortal
Q2F696
A0A1E1WHH9
A0A3S2L716
A0A212F496
A0A2W1BTQ8
A0A194R1P4
+ More
S4PGM5 H9IT18 A0A0L7KUY4 A0A0N7Z9V8 A0A194PYL8 A0A0K8TKE9 A0A182Q4W4 A0A182XZF0 A0A2M4AHP5 A0A2M4AGQ8 A0A182UV85 Q7QJV3 D6X0C9 A0A034VY92 A0A023EQA6 A0A1B0CMS5 A0A1Q3G5A1 T1E9Z3 A0A1Q3G5C2 A0A336M7P8 A0A0A1XCS0 A0A2M4CRK6 A0A2M4BSI6 A0A1L8DMA5 A0A2M4CRN3 W8BJF0 A0A2M3Z208 V5GRE7 A0A182IJL8 E9HF44 A0A084VF71 A0A0P5YJ64 A0A1S3DUX0 A0A2H1V422 A0A1Y1KAR2 A0A1L8DM09 A0A0J9R825 Q7JX82 B4HRK6 A0A0P6JS04 B4NMR5 B4P2P1 B3N8T6 A0A1J1J3W2 A0A1W4VYY5 A0A1J1J6L5 B4KNW1 B3MGT9 B4GGM9 Q28ZX6 A0A3Q0II38 A0A3Q0INK3 A0A3B0J282 T1PBJ3 A0A026W172 E2C237 A0A1B6EAC6 A0A1A9V8H2 A0A1B0FQG0 A0A1A9Z5J2 A0A1B0BV49 A0A1A9WP09 A0A1B6LBY5 B4J8C4 A0A1A9XB70 A0A158NUJ8 A0A0M5J337 B4LJN2 A0A1I8PYU9 T1I230 A0A224XSQ1 J3JVI6 A0A1B6FM06 A0A023F3Y3 A0A2H8TLD2 C4WSV7 A0A087U047 A0A146L901 A0A146KSW3 A0A0N0U608 A0A151X6L2 J9JKJ2 A0A2L2Y1Q7 A0A2S2Q945 A0A195C9C2 E2A382 A0A0P4VNI6 A0A195FIY5 C1C1V8 A0A087ZSQ2 A0A151J2S6 A0A0K2V9I6 A0A0K2V8F8 A0A2A3EEJ6
S4PGM5 H9IT18 A0A0L7KUY4 A0A0N7Z9V8 A0A194PYL8 A0A0K8TKE9 A0A182Q4W4 A0A182XZF0 A0A2M4AHP5 A0A2M4AGQ8 A0A182UV85 Q7QJV3 D6X0C9 A0A034VY92 A0A023EQA6 A0A1B0CMS5 A0A1Q3G5A1 T1E9Z3 A0A1Q3G5C2 A0A336M7P8 A0A0A1XCS0 A0A2M4CRK6 A0A2M4BSI6 A0A1L8DMA5 A0A2M4CRN3 W8BJF0 A0A2M3Z208 V5GRE7 A0A182IJL8 E9HF44 A0A084VF71 A0A0P5YJ64 A0A1S3DUX0 A0A2H1V422 A0A1Y1KAR2 A0A1L8DM09 A0A0J9R825 Q7JX82 B4HRK6 A0A0P6JS04 B4NMR5 B4P2P1 B3N8T6 A0A1J1J3W2 A0A1W4VYY5 A0A1J1J6L5 B4KNW1 B3MGT9 B4GGM9 Q28ZX6 A0A3Q0II38 A0A3Q0INK3 A0A3B0J282 T1PBJ3 A0A026W172 E2C237 A0A1B6EAC6 A0A1A9V8H2 A0A1B0FQG0 A0A1A9Z5J2 A0A1B0BV49 A0A1A9WP09 A0A1B6LBY5 B4J8C4 A0A1A9XB70 A0A158NUJ8 A0A0M5J337 B4LJN2 A0A1I8PYU9 T1I230 A0A224XSQ1 J3JVI6 A0A1B6FM06 A0A023F3Y3 A0A2H8TLD2 C4WSV7 A0A087U047 A0A146L901 A0A146KSW3 A0A0N0U608 A0A151X6L2 J9JKJ2 A0A2L2Y1Q7 A0A2S2Q945 A0A195C9C2 E2A382 A0A0P4VNI6 A0A195FIY5 C1C1V8 A0A087ZSQ2 A0A151J2S6 A0A0K2V9I6 A0A0K2V8F8 A0A2A3EEJ6
PDB
5V6T
E-value=3.87838e-86,
Score=810
Ontologies
GO
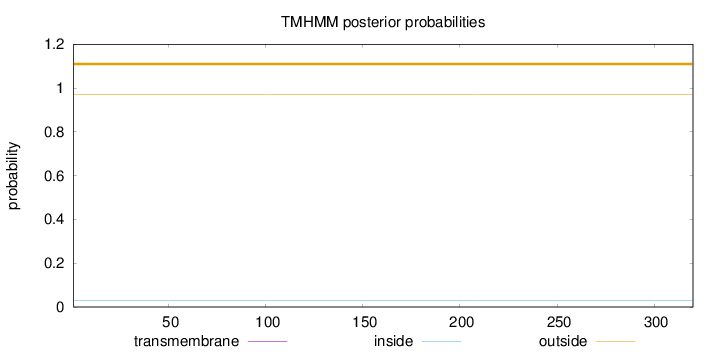
Topology
Length:
320
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000670000000000001
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.02980
outside
1 - 320
Population Genetic Test Statistics
Pi
240.590707
Theta
164.647623
Tajima's D
1.023044
CLR
0.220575
CSRT
0.667816609169542
Interpretation
Uncertain
