Gene
KWMTBOMO13070
Pre Gene Modal
BGIBMGA000399
Annotation
ribosomal_protein_L20_[Bombyx_mori]
Full name
Succinate--CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial
Alternative Name
Succinyl-CoA synthetase subunit alpha
Location in the cell
Mitochondrial Reliability : 3.052
Sequence
CDS
ATGGTATTTTTGTCTGTTATAAACTGGGCCCGCTGTCGTGGACCCGACGAATTTTGGAGAAAGCGACGGGTTTTTAGATTAGCCGCTCATTATATTGGCCGTCGACGAAATTGTTATTCGATTGCAGTTCGAAATGTTCACCGAGCTCTAGTTTATGCAACCAAAGCAAGAAAGCTTAAGAAAGAAGATATGAAAAGTCTTTGGGACGTTAGAATAACGGCTGCTTGTGAGCAACACAATATTACACTATTCTCTTTAAGAGAAGGCTTGGACAGAGCTAATATAATGTTGGATAGGAAATCTTTGTCAGACTTGGCTTCATGGGAACCAAAAACATTTGAAGCCCTCGCTGCTGTAGCTAAATACAAACTTGAAACAGACAGATTCATAGATGGACAAGATAAATGTCACCCCACAGGAATAAATCTAAACCTTAAGGATGTGATGTTAGAGGCATGGTTGAATGAAAAGAAAAAATAA
Protein
MVFLSVINWARCRGPDEFWRKRRVFRLAAHYIGRRRNCYSIAVRNVHRALVYATKARKLKKEDMKSLWDVRITAACEQHNITLFSLREGLDRANIMLDRKSLSDLASWEPKTFEALAAVAKYKLETDRFIDGQDKCHPTGINLNLKDVMLEAWLNEKKK
Summary
Description
Succinyl-CoA synthetase functions in the citric acid cycle (TCA), coupling the hydrolysis of succinyl-CoA to the synthesis of either ATP or GTP and thus represents the only step of substrate-level phosphorylation in the TCA. The alpha subunit of the enzyme binds the substrates coenzyme A and phosphate, while succinate binding and specificity for either ATP or GTP is provided by different beta subunits.
Catalytic Activity
ATP + CoA + succinate = ADP + phosphate + succinyl-CoA
CoA + GTP + succinate = GDP + phosphate + succinyl-CoA
CoA + GTP + succinate = GDP + phosphate + succinyl-CoA
Subunit
Heterodimer of an alpha and a beta subunit. Different beta subunits determine nucleotide specificity. Together with an ATP-specific beta subunit, forms an ADP-forming succinyl-CoA synthetase (A-SCS). Together with a GTP-specific beta subunit forms a GDP-forming succinyl-CoA synthetase (G-SCS).
Similarity
Belongs to the bacterial ribosomal protein bL20 family.
Belongs to the succinate/malate CoA ligase alpha subunit family.
Belongs to the succinate/malate CoA ligase alpha subunit family.
Uniprot
Q2F656
I4DKM9
A0A194R161
A0A2W1BP96
I4DMR6
Q95UN2
+ More
S4PSF2 A0A212F5V6 A0A1B0CUR3 A0A1L8DR99 A0A1B0DAD7 A0A1Y1LP30 A0A1I8JV36 A0A240PM15 A0A2M4C3S3 A0A1Y9G8G2 A0A2M4C435 A0A1I8JVJ7 A0A1I8JTD1 A0A1C7CZS8 A7UVP7 A0A2M4C439 T1DH43 A0A1I8JW46 A0A2M4AR59 A0A2M4AR72 W5J4X0 A0A0P4VH45 A0A0V0G6H5 A0A240PLH4 Q16HN7 A0A2M3ZJR6 U5EGM8 A0A240PLQ6 A0A2M3ZCU7 A0A224XN64 A0A2M3ZK04 A0A1W4WHU5 A0A2C9GUJ9 A0A2M3ZJU0 A0A182YBD5 A0A1Y9HB34 A0A182SV10 N6UP67 A0A067R4B4 D6WJ74 A0A1Y9IV39 A0A084VXV8 A0A1Y9H380 A0A3F2YXL7 A0A023EGF5 J3JXD0 V5IAL4 A0A0A9XTS5 A0A2J7QKQ2 A0A1Q3FCQ1 A0A1L8EDN1 A0A1L8EAH9 R4WQB3 A0A0L7RD94 B0WSS3 A0A088ARQ4 A0A195F2Z5 A0A1I8PME7 A0A0K8TPB0 A0A2A3E2F6 A0A182GIX5 A0A182G411 A0A195DCE5 A0A158P311 A0A2R7X3F3 A0A0L0C3Y1 A0A195BCZ5 W8C256 A0A1A9WH21 A0A1A9Z1R0 A0A0K8VUZ0 A0A0M8ZM77 A0A0A1X1G4 A0A1A9Y9U9 A0A1B0BWM3 A0A1I8NHC3 E9IEY1 A0A1B0FA11 A0A3B0J411 A0A1A9WCT2 Q2M0C2 B4GRU2 A0A1W4VIX8 B4LBH4 B4QIY3 A0A154PA40 B4HGN3 Q9VU36 B4ML22 A0A1A9VMQ8 B4PHB1 A0A1B6C6W6 B3MAU3
S4PSF2 A0A212F5V6 A0A1B0CUR3 A0A1L8DR99 A0A1B0DAD7 A0A1Y1LP30 A0A1I8JV36 A0A240PM15 A0A2M4C3S3 A0A1Y9G8G2 A0A2M4C435 A0A1I8JVJ7 A0A1I8JTD1 A0A1C7CZS8 A7UVP7 A0A2M4C439 T1DH43 A0A1I8JW46 A0A2M4AR59 A0A2M4AR72 W5J4X0 A0A0P4VH45 A0A0V0G6H5 A0A240PLH4 Q16HN7 A0A2M3ZJR6 U5EGM8 A0A240PLQ6 A0A2M3ZCU7 A0A224XN64 A0A2M3ZK04 A0A1W4WHU5 A0A2C9GUJ9 A0A2M3ZJU0 A0A182YBD5 A0A1Y9HB34 A0A182SV10 N6UP67 A0A067R4B4 D6WJ74 A0A1Y9IV39 A0A084VXV8 A0A1Y9H380 A0A3F2YXL7 A0A023EGF5 J3JXD0 V5IAL4 A0A0A9XTS5 A0A2J7QKQ2 A0A1Q3FCQ1 A0A1L8EDN1 A0A1L8EAH9 R4WQB3 A0A0L7RD94 B0WSS3 A0A088ARQ4 A0A195F2Z5 A0A1I8PME7 A0A0K8TPB0 A0A2A3E2F6 A0A182GIX5 A0A182G411 A0A195DCE5 A0A158P311 A0A2R7X3F3 A0A0L0C3Y1 A0A195BCZ5 W8C256 A0A1A9WH21 A0A1A9Z1R0 A0A0K8VUZ0 A0A0M8ZM77 A0A0A1X1G4 A0A1A9Y9U9 A0A1B0BWM3 A0A1I8NHC3 E9IEY1 A0A1B0FA11 A0A3B0J411 A0A1A9WCT2 Q2M0C2 B4GRU2 A0A1W4VIX8 B4LBH4 B4QIY3 A0A154PA40 B4HGN3 Q9VU36 B4ML22 A0A1A9VMQ8 B4PHB1 A0A1B6C6W6 B3MAU3
EC Number
6.2.1.4
6.2.1.5
6.2.1.5
Pubmed
19121390
22651552
26354079
28756777
23622113
22118469
+ More
28004739 20966253 12364791 20920257 23761445 27129103 17510324 25244985 23537049 24845553 18362917 19820115 24438588 24945155 22516182 25401762 26823975 23691247 26369729 26483478 21347285 26108605 24495485 25830018 25315136 21282665 15632085 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
28004739 20966253 12364791 20920257 23761445 27129103 17510324 25244985 23537049 24845553 18362917 19820115 24438588 24945155 22516182 25401762 26823975 23691247 26369729 26483478 21347285 26108605 24495485 25830018 25315136 21282665 15632085 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
EMBL
BABH01013186
DQ311216
ABD36161.1
AK401847
KQ459586
BAM18469.1
+ More
KPI97841.1 KQ460883 KPJ11442.1 KZ150009 PZC75127.1 AK402584 BAM19206.1 AY046535 AAL03954.1 GAIX01013893 JAA78667.1 AGBW02010128 OWR49116.1 AJWK01029612 GFDF01005092 JAV08992.1 AJVK01013276 GEZM01051805 JAV74691.1 GGFJ01010680 MBW59821.1 GGFJ01010770 MBW59911.1 APCN01000834 AAAB01008987 EDO63271.2 GGFJ01010627 MBW59768.1 GAMD01002506 JAA99084.1 GGFK01009881 MBW43202.1 GGFK01009953 MBW43274.1 ADMH02002105 ETN59001.1 GDKW01002870 JAI53725.1 GECL01002425 JAP03699.1 CH478156 EAT33765.1 GGFM01007991 MBW28742.1 GANO01003380 JAB56491.1 GGFM01005623 MBW26374.1 GFTR01002490 JAW13936.1 GGFM01008064 MBW28815.1 AXCM01001181 GGFM01008021 MBW28772.1 AXCN02001518 APGK01023663 KB740510 ENN80522.1 KK852902 KDR14058.1 KQ971343 EFA03160.1 ATLV01018208 KE525224 KFB42802.1 GAPW01005874 JAC07724.1 BT127899 KB632104 AEE62861.1 ERL88863.1 GALX01000980 JAB67486.1 GBHO01021376 GBRD01015877 GDHC01008808 JAG22228.1 JAG49949.1 JAQ09821.1 NEVH01013262 PNF29149.1 GFDL01009690 JAV25355.1 GFDG01001978 JAV16821.1 GFDG01003068 JAV15731.1 AK417886 BAN21101.1 KQ414614 KOC68828.1 DS232075 EDS34032.1 KQ981855 KYN34756.1 GDAI01001622 JAI15981.1 KZ288427 PBC25878.1 JXUM01067006 JXUM01067007 JXUM01067008 KQ562428 KXJ75934.1 JXUM01041704 JXUM01041705 KQ561294 KXJ79084.1 KQ980989 KYN10561.1 ADTU01001303 KK856302 PTY25435.1 JRES01000945 KNC26936.1 KQ976522 KYM82082.1 GAMC01000643 JAC05913.1 GDHF01009607 JAI42707.1 KQ438551 KOX67170.1 GBXI01009576 JAD04716.1 JXJN01021871 GL762728 EFZ20868.1 CCAG010018114 OUUW01000002 SPP76167.1 CH379069 EAL31012.1 CH479188 EDW40477.1 CH940647 EDW70784.1 CM000363 CM002912 EDX10314.1 KMY99348.1 KQ434856 KZC08703.1 CH480815 EDW41341.1 AE014296 BT089020 AAF49855.1 ACT98667.1 CH963847 EDW73080.1 CM000159 EDW94372.1 GEDC01028052 GEDC01007332 JAS09246.1 JAS29966.1 CH902618 EDV39177.1
KPI97841.1 KQ460883 KPJ11442.1 KZ150009 PZC75127.1 AK402584 BAM19206.1 AY046535 AAL03954.1 GAIX01013893 JAA78667.1 AGBW02010128 OWR49116.1 AJWK01029612 GFDF01005092 JAV08992.1 AJVK01013276 GEZM01051805 JAV74691.1 GGFJ01010680 MBW59821.1 GGFJ01010770 MBW59911.1 APCN01000834 AAAB01008987 EDO63271.2 GGFJ01010627 MBW59768.1 GAMD01002506 JAA99084.1 GGFK01009881 MBW43202.1 GGFK01009953 MBW43274.1 ADMH02002105 ETN59001.1 GDKW01002870 JAI53725.1 GECL01002425 JAP03699.1 CH478156 EAT33765.1 GGFM01007991 MBW28742.1 GANO01003380 JAB56491.1 GGFM01005623 MBW26374.1 GFTR01002490 JAW13936.1 GGFM01008064 MBW28815.1 AXCM01001181 GGFM01008021 MBW28772.1 AXCN02001518 APGK01023663 KB740510 ENN80522.1 KK852902 KDR14058.1 KQ971343 EFA03160.1 ATLV01018208 KE525224 KFB42802.1 GAPW01005874 JAC07724.1 BT127899 KB632104 AEE62861.1 ERL88863.1 GALX01000980 JAB67486.1 GBHO01021376 GBRD01015877 GDHC01008808 JAG22228.1 JAG49949.1 JAQ09821.1 NEVH01013262 PNF29149.1 GFDL01009690 JAV25355.1 GFDG01001978 JAV16821.1 GFDG01003068 JAV15731.1 AK417886 BAN21101.1 KQ414614 KOC68828.1 DS232075 EDS34032.1 KQ981855 KYN34756.1 GDAI01001622 JAI15981.1 KZ288427 PBC25878.1 JXUM01067006 JXUM01067007 JXUM01067008 KQ562428 KXJ75934.1 JXUM01041704 JXUM01041705 KQ561294 KXJ79084.1 KQ980989 KYN10561.1 ADTU01001303 KK856302 PTY25435.1 JRES01000945 KNC26936.1 KQ976522 KYM82082.1 GAMC01000643 JAC05913.1 GDHF01009607 JAI42707.1 KQ438551 KOX67170.1 GBXI01009576 JAD04716.1 JXJN01021871 GL762728 EFZ20868.1 CCAG010018114 OUUW01000002 SPP76167.1 CH379069 EAL31012.1 CH479188 EDW40477.1 CH940647 EDW70784.1 CM000363 CM002912 EDX10314.1 KMY99348.1 KQ434856 KZC08703.1 CH480815 EDW41341.1 AE014296 BT089020 AAF49855.1 ACT98667.1 CH963847 EDW73080.1 CM000159 EDW94372.1 GEDC01028052 GEDC01007332 JAS09246.1 JAS29966.1 CH902618 EDV39177.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000092461
UP000092462
+ More
UP000075902 UP000075885 UP000069272 UP000075903 UP000075840 UP000075882 UP000007062 UP000076407 UP000000673 UP000075880 UP000008820 UP000075881 UP000192223 UP000075883 UP000076408 UP000075886 UP000075901 UP000019118 UP000027135 UP000007266 UP000075920 UP000030765 UP000075884 UP000075900 UP000030742 UP000235965 UP000053825 UP000002320 UP000005203 UP000078541 UP000095300 UP000242457 UP000069940 UP000249989 UP000078492 UP000005205 UP000037069 UP000078540 UP000091820 UP000092445 UP000053105 UP000092443 UP000092460 UP000095301 UP000092444 UP000268350 UP000001819 UP000008744 UP000192221 UP000008792 UP000000304 UP000076502 UP000001292 UP000000803 UP000007798 UP000078200 UP000002282 UP000007801
UP000075902 UP000075885 UP000069272 UP000075903 UP000075840 UP000075882 UP000007062 UP000076407 UP000000673 UP000075880 UP000008820 UP000075881 UP000192223 UP000075883 UP000076408 UP000075886 UP000075901 UP000019118 UP000027135 UP000007266 UP000075920 UP000030765 UP000075884 UP000075900 UP000030742 UP000235965 UP000053825 UP000002320 UP000005203 UP000078541 UP000095300 UP000242457 UP000069940 UP000249989 UP000078492 UP000005205 UP000037069 UP000078540 UP000091820 UP000092445 UP000053105 UP000092443 UP000092460 UP000095301 UP000092444 UP000268350 UP000001819 UP000008744 UP000192221 UP000008792 UP000000304 UP000076502 UP000001292 UP000000803 UP000007798 UP000078200 UP000002282 UP000007801
Pfam
Interpro
IPR005813
Ribosomal_L20
+ More
IPR035566 Ribosomal_protein_L20_C
IPR024135 LAMTOR5
IPR036291 NAD(P)-bd_dom_sf
IPR005811 CoA_ligase
IPR003781 CoA-bd
IPR033847 Citrt_syn/SCS-alpha_CS
IPR005810 CoA_lig_alpha
IPR016102 Succinyl-CoA_synth-like
IPR017440 Cit_synth/succinyl-CoA_lig_AS
IPR029045 ClpP/crotonase-like_dom_sf
IPR034733 AcCoA_carboxyl
IPR011762 COA_CT_N
IPR011763 COA_CT_C
IPR035566 Ribosomal_protein_L20_C
IPR024135 LAMTOR5
IPR036291 NAD(P)-bd_dom_sf
IPR005811 CoA_ligase
IPR003781 CoA-bd
IPR033847 Citrt_syn/SCS-alpha_CS
IPR005810 CoA_lig_alpha
IPR016102 Succinyl-CoA_synth-like
IPR017440 Cit_synth/succinyl-CoA_lig_AS
IPR029045 ClpP/crotonase-like_dom_sf
IPR034733 AcCoA_carboxyl
IPR011762 COA_CT_N
IPR011763 COA_CT_C
Gene 3D
ProteinModelPortal
Q2F656
I4DKM9
A0A194R161
A0A2W1BP96
I4DMR6
Q95UN2
+ More
S4PSF2 A0A212F5V6 A0A1B0CUR3 A0A1L8DR99 A0A1B0DAD7 A0A1Y1LP30 A0A1I8JV36 A0A240PM15 A0A2M4C3S3 A0A1Y9G8G2 A0A2M4C435 A0A1I8JVJ7 A0A1I8JTD1 A0A1C7CZS8 A7UVP7 A0A2M4C439 T1DH43 A0A1I8JW46 A0A2M4AR59 A0A2M4AR72 W5J4X0 A0A0P4VH45 A0A0V0G6H5 A0A240PLH4 Q16HN7 A0A2M3ZJR6 U5EGM8 A0A240PLQ6 A0A2M3ZCU7 A0A224XN64 A0A2M3ZK04 A0A1W4WHU5 A0A2C9GUJ9 A0A2M3ZJU0 A0A182YBD5 A0A1Y9HB34 A0A182SV10 N6UP67 A0A067R4B4 D6WJ74 A0A1Y9IV39 A0A084VXV8 A0A1Y9H380 A0A3F2YXL7 A0A023EGF5 J3JXD0 V5IAL4 A0A0A9XTS5 A0A2J7QKQ2 A0A1Q3FCQ1 A0A1L8EDN1 A0A1L8EAH9 R4WQB3 A0A0L7RD94 B0WSS3 A0A088ARQ4 A0A195F2Z5 A0A1I8PME7 A0A0K8TPB0 A0A2A3E2F6 A0A182GIX5 A0A182G411 A0A195DCE5 A0A158P311 A0A2R7X3F3 A0A0L0C3Y1 A0A195BCZ5 W8C256 A0A1A9WH21 A0A1A9Z1R0 A0A0K8VUZ0 A0A0M8ZM77 A0A0A1X1G4 A0A1A9Y9U9 A0A1B0BWM3 A0A1I8NHC3 E9IEY1 A0A1B0FA11 A0A3B0J411 A0A1A9WCT2 Q2M0C2 B4GRU2 A0A1W4VIX8 B4LBH4 B4QIY3 A0A154PA40 B4HGN3 Q9VU36 B4ML22 A0A1A9VMQ8 B4PHB1 A0A1B6C6W6 B3MAU3
S4PSF2 A0A212F5V6 A0A1B0CUR3 A0A1L8DR99 A0A1B0DAD7 A0A1Y1LP30 A0A1I8JV36 A0A240PM15 A0A2M4C3S3 A0A1Y9G8G2 A0A2M4C435 A0A1I8JVJ7 A0A1I8JTD1 A0A1C7CZS8 A7UVP7 A0A2M4C439 T1DH43 A0A1I8JW46 A0A2M4AR59 A0A2M4AR72 W5J4X0 A0A0P4VH45 A0A0V0G6H5 A0A240PLH4 Q16HN7 A0A2M3ZJR6 U5EGM8 A0A240PLQ6 A0A2M3ZCU7 A0A224XN64 A0A2M3ZK04 A0A1W4WHU5 A0A2C9GUJ9 A0A2M3ZJU0 A0A182YBD5 A0A1Y9HB34 A0A182SV10 N6UP67 A0A067R4B4 D6WJ74 A0A1Y9IV39 A0A084VXV8 A0A1Y9H380 A0A3F2YXL7 A0A023EGF5 J3JXD0 V5IAL4 A0A0A9XTS5 A0A2J7QKQ2 A0A1Q3FCQ1 A0A1L8EDN1 A0A1L8EAH9 R4WQB3 A0A0L7RD94 B0WSS3 A0A088ARQ4 A0A195F2Z5 A0A1I8PME7 A0A0K8TPB0 A0A2A3E2F6 A0A182GIX5 A0A182G411 A0A195DCE5 A0A158P311 A0A2R7X3F3 A0A0L0C3Y1 A0A195BCZ5 W8C256 A0A1A9WH21 A0A1A9Z1R0 A0A0K8VUZ0 A0A0M8ZM77 A0A0A1X1G4 A0A1A9Y9U9 A0A1B0BWM3 A0A1I8NHC3 E9IEY1 A0A1B0FA11 A0A3B0J411 A0A1A9WCT2 Q2M0C2 B4GRU2 A0A1W4VIX8 B4LBH4 B4QIY3 A0A154PA40 B4HGN3 Q9VU36 B4ML22 A0A1A9VMQ8 B4PHB1 A0A1B6C6W6 B3MAU3
PDB
6GB2
E-value=6.5811e-24,
Score=268
Ontologies
GO
PANTHER
Topology
Subcellular location
Mitochondrion
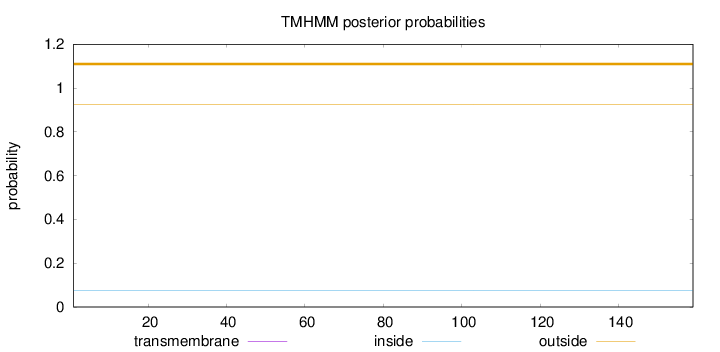
Length:
159
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000790000000000001
Exp number, first 60 AAs:
0.00037
Total prob of N-in:
0.07528
outside
1 - 159
Population Genetic Test Statistics
Pi
222.257428
Theta
185.113309
Tajima's D
0.498932
CLR
0.072297
CSRT
0.516974151292435
Interpretation
Uncertain
