Gene
KWMTBOMO13068
Pre Gene Modal
BGIBMGA000301
Annotation
PREDICTED:_pancreatic_triacylglycerol_lipase-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.089 Extracellular Reliability : 1.187 Mitochondrial Reliability : 1.132
Sequence
CDS
ATGCGCAGACTTCACCTCACAGACAGACATCTTCGAGTTATGCTGTGGTGTATCCTGTTAGGTCTGCAGCCAGCGTTTGGTGGAATACTTGACCCCTTCAACTGGGCAAGATCAGATAGAATCGAAGTCAACATACCATGGCTACCATTTGAAAATGAAACAAGATGCTATGATGAACTGGGTTGTCTCAACATTACAAGGAGCTGGTATCACCTGATTCATCGTCCCTTCAACGTGTTTCCGTTACCACGAGTTGTCATCAATACCAGGTTCATTTTGTATACAAAGAAGAACCCGACTGATGGTCAACTACTGAACGTGAATGTGAATAAGACGATAGTGAAATCGAATTTCGACCCTCAGAAACCTACGAAGATGATCGTACACGGATTCATCGATACACCGCTTTCTAATTGGGTCAGTGAAATGAAAGACGAACTCGTAAAAGCCGGTGACTTTAACGTTGTTATTGTGGACTGGGCGGGAGGGAGTCTACCGCTCTACACCCAGGCTACAGCAAATACAAGGCTTGTGGGTTTAGAAATCGCTCATTTTATCAATACTTTAGAGAAAGACCACGGATTAAATCCTCAAGATGTCCACATAATAGGACACTCTTTAGGTGCTCACACGGCTGGTTACGCTGGAGAACGTATACAAGGTTTAGGAAGAATAACGGGTCTCGATCCAGCAGAACCTTACTTTCAAGGAATGCCAACACACATCAGGTTGGATCCAACGGATGCTCAATTGGTCGATGTCATACATACAGATGGAAAGAGCATTTTCCTTTTAGGATACGGAATGTCGCAGCCAGTGGGCCATTTAGATTTTTATCCTAATAATGGTAAAGAACAACCAGGCTGCGATCTCACAGAGGGGCCACTGGTGCCGCTAACTCTAGTCAAGCAGGGCCTTGAGGAAGCTTCAAGGGTTTTAGTAGCCTGCAACCATGTTAGAGCTATAAAATTGTTTACAGAATCGATCAATGGAAAATGTGCCTATATAGGGCATCAATGCCCATCGTACCAGCATTTTTTGGCAGGAAAATGCTTCCACTGTGGTCATGGTTGCGCTGTTATGGGTTTCCATGCAGACAGTACTCCAGGACTAGTAGCGAACAGGGACAATTTGACAGAAAACGAGGTATACCCATCTGAAGAACATGAAACCATTGGGGCTAAATACTTCCTCAGTACCGGCAAAGAACAACCGTTTTGCCAGAGACACTATAGAGTGACCATTCAACTAGCAAATCCTAAAGGAGCTGAATCTTGGGTCCAGGGGTTTTTGAAGGTAGCACTTTTATCTGACCGTGGAGTAATTAGGGGTATGGACCTAACACCAAACAACTATGTAAAGTTAGAACACGGCAGCACTTACACCATTGTTGTTACGCATCCAATGGATATGGGTGGTAAAGTTAGGAAGGTGGAGTTATCGTGGGAGTACGACATGAATGTACTAGAGCCGCGATCCCTTTGCATCCTGTGGTGTAATGATCGATTATATGTCAAATCTGTCATTGTTGATCAAATGGAATTACCATCGAGAGGGAAAAGAAATGTTGATTTTAGTAGTAAATTGTGTACACTAAAAAGAGAATTCGCCGAGATACCCAATAGAGGTTCTGCAAGCTTCTATGACAACTGCGGGAGGTAG
Protein
MRRLHLTDRHLRVMLWCILLGLQPAFGGILDPFNWARSDRIEVNIPWLPFENETRCYDELGCLNITRSWYHLIHRPFNVFPLPRVVINTRFILYTKKNPTDGQLLNVNVNKTIVKSNFDPQKPTKMIVHGFIDTPLSNWVSEMKDELVKAGDFNVVIVDWAGGSLPLYTQATANTRLVGLEIAHFINTLEKDHGLNPQDVHIIGHSLGAHTAGYAGERIQGLGRITGLDPAEPYFQGMPTHIRLDPTDAQLVDVIHTDGKSIFLLGYGMSQPVGHLDFYPNNGKEQPGCDLTEGPLVPLTLVKQGLEEASRVLVACNHVRAIKLFTESINGKCAYIGHQCPSYQHFLAGKCFHCGHGCAVMGFHADSTPGLVANRDNLTENEVYPSEEHETIGAKYFLSTGKEQPFCQRHYRVTIQLANPKGAESWVQGFLKVALLSDRGVIRGMDLTPNNYVKLEHGSTYTIVVTHPMDMGGKVRKVELSWEYDMNVLEPRSLCILWCNDRLYVKSVIVDQMELPSRGKRNVDFSSKLCTLKREFAEIPNRGSASFYDNCGR
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
H9ISS4
A0A2W1BP69
A0A194R1P9
A0A194PYL3
A0A212F5W2
A0A2H1V686
+ More
A0A0L7R0F6 A0A158NHG2 A0A154P9H3 A0A0C9RBL4 A0A195D4C8 A0A195AVA3 K7J6W4 A0A3L8DIF3 A0A026WTB1 A0A0M8ZPG5 A0A1Y1KW87 A0A2A3E1P8 E1ZYD7 A0A232EW00 A0A1W4XTY6 A0A2J7RLM3 A0A182V3L0 A0A139WC96 A0A182KRZ0 A0A336L9N3 A0A182UET5 A0A182X6I3 Q7QJV2 A0A182I3T4 A0A182QN63 A0A182NHF4 A0A182XZF1 A0A182P9X9 A0A182WIV7 A0A182RB46 A0A182LTP7 A0A084VF72 A0A182FCR9 A0A182JQV4 A0A0G3YJ76 J9JX72 A0A2H8TRX0 A0A182JG74 A0A182GWE3 F4WDB3 T1H7M9 A0A1B6K1C3 A0A1B0CMS4 A0A224XHR8 A0A195FW40 A0A2H8TER6 A0A1S3CYD5 A0A195E2C6 A0A1S4FX83 Q16LG0 A0A151XFM1 E0VGW7 A0A0A9WBI9 A0A1J1J3X0 A0A0K8STE1 A0A1L4AAS6 B0VZR7 E2C2J9 A0A2J7RLQ3 A0A2P8Y5U6 A0A2R7WV01 W5JIZ1 A0A2J7RLM4 A0A336MRK7 A0A1B6DBF2 A0A1B6F8P8 A0A1D2MJV0 A0A0P5J9I4 A0A164PW02 A0A0P6BIX5 A0A0P5K9M7 A0A0P6GY14 A0A226E6A6 A0A0N8CTH7 E9H0C7 A0A323TKN3 A0A3R7PD87 A0A0P5J225 A0A2S1J5N0 A0A0P5NYM5 V4A6S5
A0A0L7R0F6 A0A158NHG2 A0A154P9H3 A0A0C9RBL4 A0A195D4C8 A0A195AVA3 K7J6W4 A0A3L8DIF3 A0A026WTB1 A0A0M8ZPG5 A0A1Y1KW87 A0A2A3E1P8 E1ZYD7 A0A232EW00 A0A1W4XTY6 A0A2J7RLM3 A0A182V3L0 A0A139WC96 A0A182KRZ0 A0A336L9N3 A0A182UET5 A0A182X6I3 Q7QJV2 A0A182I3T4 A0A182QN63 A0A182NHF4 A0A182XZF1 A0A182P9X9 A0A182WIV7 A0A182RB46 A0A182LTP7 A0A084VF72 A0A182FCR9 A0A182JQV4 A0A0G3YJ76 J9JX72 A0A2H8TRX0 A0A182JG74 A0A182GWE3 F4WDB3 T1H7M9 A0A1B6K1C3 A0A1B0CMS4 A0A224XHR8 A0A195FW40 A0A2H8TER6 A0A1S3CYD5 A0A195E2C6 A0A1S4FX83 Q16LG0 A0A151XFM1 E0VGW7 A0A0A9WBI9 A0A1J1J3X0 A0A0K8STE1 A0A1L4AAS6 B0VZR7 E2C2J9 A0A2J7RLQ3 A0A2P8Y5U6 A0A2R7WV01 W5JIZ1 A0A2J7RLM4 A0A336MRK7 A0A1B6DBF2 A0A1B6F8P8 A0A1D2MJV0 A0A0P5J9I4 A0A164PW02 A0A0P6BIX5 A0A0P5K9M7 A0A0P6GY14 A0A226E6A6 A0A0N8CTH7 E9H0C7 A0A323TKN3 A0A3R7PD87 A0A0P5J225 A0A2S1J5N0 A0A0P5NYM5 V4A6S5
Pubmed
EMBL
BABH01013187
KZ150009
PZC75097.1
KQ460883
KPJ11444.1
KQ459586
+ More
KPI97839.1 AGBW02010128 OWR49114.1 ODYU01000702 SOQ35912.1 KQ414670 KOC64286.1 ADTU01015802 ADTU01015803 KQ434832 KZC07878.1 GBYB01013914 JAG83681.1 KQ976870 KYN07755.1 KQ976737 KYM75904.1 AAZX01009912 QOIP01000008 RLU19618.1 KK107109 EZA59183.1 KQ436240 KOX67326.1 GEZM01073773 JAV64838.1 KZ288481 PBC25252.1 GL435204 EFN73836.1 NNAY01001937 OXU22506.1 NEVH01002684 PNF41733.1 KQ971370 KYB25598.1 UFQS01002488 UFQT01002488 SSX14149.1 SSX33565.1 AAAB01008807 EAA04199.5 APCN01000220 AXCN02000591 AXCM01000560 ATLV01012364 KE524785 KFB36616.1 KM507111 AKM28435.1 ABLF02025583 GFXV01004537 MBW16342.1 JXUM01092976 JXUM01092977 KQ564025 KXJ72902.1 GL888086 EGI67840.1 ACPB03000883 GECU01002462 JAT05245.1 AJWK01019143 GFTR01007078 JAW09348.1 KQ981208 KYN44875.1 GFXV01000792 GFXV01007163 MBW12597.1 MBW18968.1 KQ979763 KYN19231.1 CH477907 EAT35159.1 KQ982194 KYQ59068.1 DS235152 EEB12623.1 GBHO01041389 JAG02215.1 CVRI01000070 CRL07123.1 GBRD01009273 JAG56548.1 KX061494 API65149.1 DS231815 EDS35159.1 GL452135 EFN77855.1 PNF41729.1 PYGN01000894 PSN39632.1 KK855638 PTY23378.1 ADMH02001104 ETN64101.1 PNF41730.1 UFQT01001685 SSX31423.1 GEDC01014284 JAS23014.1 GECZ01023157 GECZ01000844 JAS46612.1 JAS68925.1 LJIJ01001029 ODM93289.1 GDIQ01204753 JAK46972.1 LRGB01002544 KZS07195.1 GDIP01014378 JAM89337.1 GDIQ01187452 JAK64273.1 GDIQ01027662 JAN67075.1 LNIX01000007 OXA52096.1 GDIP01100431 JAM03284.1 GL732580 EFX74783.1 MDVM02000038 PYZ99352.1 QCYY01002831 ROT67232.1 GDIQ01206209 JAK45516.1 MF182405 AWF73664.1 GDIQ01137981 JAL13745.1 KB202094 ESO92407.1
KPI97839.1 AGBW02010128 OWR49114.1 ODYU01000702 SOQ35912.1 KQ414670 KOC64286.1 ADTU01015802 ADTU01015803 KQ434832 KZC07878.1 GBYB01013914 JAG83681.1 KQ976870 KYN07755.1 KQ976737 KYM75904.1 AAZX01009912 QOIP01000008 RLU19618.1 KK107109 EZA59183.1 KQ436240 KOX67326.1 GEZM01073773 JAV64838.1 KZ288481 PBC25252.1 GL435204 EFN73836.1 NNAY01001937 OXU22506.1 NEVH01002684 PNF41733.1 KQ971370 KYB25598.1 UFQS01002488 UFQT01002488 SSX14149.1 SSX33565.1 AAAB01008807 EAA04199.5 APCN01000220 AXCN02000591 AXCM01000560 ATLV01012364 KE524785 KFB36616.1 KM507111 AKM28435.1 ABLF02025583 GFXV01004537 MBW16342.1 JXUM01092976 JXUM01092977 KQ564025 KXJ72902.1 GL888086 EGI67840.1 ACPB03000883 GECU01002462 JAT05245.1 AJWK01019143 GFTR01007078 JAW09348.1 KQ981208 KYN44875.1 GFXV01000792 GFXV01007163 MBW12597.1 MBW18968.1 KQ979763 KYN19231.1 CH477907 EAT35159.1 KQ982194 KYQ59068.1 DS235152 EEB12623.1 GBHO01041389 JAG02215.1 CVRI01000070 CRL07123.1 GBRD01009273 JAG56548.1 KX061494 API65149.1 DS231815 EDS35159.1 GL452135 EFN77855.1 PNF41729.1 PYGN01000894 PSN39632.1 KK855638 PTY23378.1 ADMH02001104 ETN64101.1 PNF41730.1 UFQT01001685 SSX31423.1 GEDC01014284 JAS23014.1 GECZ01023157 GECZ01000844 JAS46612.1 JAS68925.1 LJIJ01001029 ODM93289.1 GDIQ01204753 JAK46972.1 LRGB01002544 KZS07195.1 GDIP01014378 JAM89337.1 GDIQ01187452 JAK64273.1 GDIQ01027662 JAN67075.1 LNIX01000007 OXA52096.1 GDIP01100431 JAM03284.1 GL732580 EFX74783.1 MDVM02000038 PYZ99352.1 QCYY01002831 ROT67232.1 GDIQ01206209 JAK45516.1 MF182405 AWF73664.1 GDIQ01137981 JAL13745.1 KB202094 ESO92407.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000053825
UP000005205
+ More
UP000076502 UP000078542 UP000078540 UP000002358 UP000279307 UP000053097 UP000053105 UP000242457 UP000000311 UP000215335 UP000192223 UP000235965 UP000075903 UP000007266 UP000075882 UP000075902 UP000076407 UP000007062 UP000075840 UP000075886 UP000075884 UP000076408 UP000075885 UP000075920 UP000075900 UP000075883 UP000030765 UP000069272 UP000075881 UP000007819 UP000075880 UP000069940 UP000249989 UP000007755 UP000015103 UP000092461 UP000078541 UP000079169 UP000078492 UP000008820 UP000075809 UP000009046 UP000183832 UP000002320 UP000008237 UP000245037 UP000000673 UP000094527 UP000076858 UP000198287 UP000000305 UP000247427 UP000283509 UP000030746
UP000076502 UP000078542 UP000078540 UP000002358 UP000279307 UP000053097 UP000053105 UP000242457 UP000000311 UP000215335 UP000192223 UP000235965 UP000075903 UP000007266 UP000075882 UP000075902 UP000076407 UP000007062 UP000075840 UP000075886 UP000075884 UP000076408 UP000075885 UP000075920 UP000075900 UP000075883 UP000030765 UP000069272 UP000075881 UP000007819 UP000075880 UP000069940 UP000249989 UP000007755 UP000015103 UP000092461 UP000078541 UP000079169 UP000078492 UP000008820 UP000075809 UP000009046 UP000183832 UP000002320 UP000008237 UP000245037 UP000000673 UP000094527 UP000076858 UP000198287 UP000000305 UP000247427 UP000283509 UP000030746
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9ISS4
A0A2W1BP69
A0A194R1P9
A0A194PYL3
A0A212F5W2
A0A2H1V686
+ More
A0A0L7R0F6 A0A158NHG2 A0A154P9H3 A0A0C9RBL4 A0A195D4C8 A0A195AVA3 K7J6W4 A0A3L8DIF3 A0A026WTB1 A0A0M8ZPG5 A0A1Y1KW87 A0A2A3E1P8 E1ZYD7 A0A232EW00 A0A1W4XTY6 A0A2J7RLM3 A0A182V3L0 A0A139WC96 A0A182KRZ0 A0A336L9N3 A0A182UET5 A0A182X6I3 Q7QJV2 A0A182I3T4 A0A182QN63 A0A182NHF4 A0A182XZF1 A0A182P9X9 A0A182WIV7 A0A182RB46 A0A182LTP7 A0A084VF72 A0A182FCR9 A0A182JQV4 A0A0G3YJ76 J9JX72 A0A2H8TRX0 A0A182JG74 A0A182GWE3 F4WDB3 T1H7M9 A0A1B6K1C3 A0A1B0CMS4 A0A224XHR8 A0A195FW40 A0A2H8TER6 A0A1S3CYD5 A0A195E2C6 A0A1S4FX83 Q16LG0 A0A151XFM1 E0VGW7 A0A0A9WBI9 A0A1J1J3X0 A0A0K8STE1 A0A1L4AAS6 B0VZR7 E2C2J9 A0A2J7RLQ3 A0A2P8Y5U6 A0A2R7WV01 W5JIZ1 A0A2J7RLM4 A0A336MRK7 A0A1B6DBF2 A0A1B6F8P8 A0A1D2MJV0 A0A0P5J9I4 A0A164PW02 A0A0P6BIX5 A0A0P5K9M7 A0A0P6GY14 A0A226E6A6 A0A0N8CTH7 E9H0C7 A0A323TKN3 A0A3R7PD87 A0A0P5J225 A0A2S1J5N0 A0A0P5NYM5 V4A6S5
A0A0L7R0F6 A0A158NHG2 A0A154P9H3 A0A0C9RBL4 A0A195D4C8 A0A195AVA3 K7J6W4 A0A3L8DIF3 A0A026WTB1 A0A0M8ZPG5 A0A1Y1KW87 A0A2A3E1P8 E1ZYD7 A0A232EW00 A0A1W4XTY6 A0A2J7RLM3 A0A182V3L0 A0A139WC96 A0A182KRZ0 A0A336L9N3 A0A182UET5 A0A182X6I3 Q7QJV2 A0A182I3T4 A0A182QN63 A0A182NHF4 A0A182XZF1 A0A182P9X9 A0A182WIV7 A0A182RB46 A0A182LTP7 A0A084VF72 A0A182FCR9 A0A182JQV4 A0A0G3YJ76 J9JX72 A0A2H8TRX0 A0A182JG74 A0A182GWE3 F4WDB3 T1H7M9 A0A1B6K1C3 A0A1B0CMS4 A0A224XHR8 A0A195FW40 A0A2H8TER6 A0A1S3CYD5 A0A195E2C6 A0A1S4FX83 Q16LG0 A0A151XFM1 E0VGW7 A0A0A9WBI9 A0A1J1J3X0 A0A0K8STE1 A0A1L4AAS6 B0VZR7 E2C2J9 A0A2J7RLQ3 A0A2P8Y5U6 A0A2R7WV01 W5JIZ1 A0A2J7RLM4 A0A336MRK7 A0A1B6DBF2 A0A1B6F8P8 A0A1D2MJV0 A0A0P5J9I4 A0A164PW02 A0A0P6BIX5 A0A0P5K9M7 A0A0P6GY14 A0A226E6A6 A0A0N8CTH7 E9H0C7 A0A323TKN3 A0A3R7PD87 A0A0P5J225 A0A2S1J5N0 A0A0P5NYM5 V4A6S5
PDB
1N8S
E-value=6.57446e-86,
Score=810
Ontologies
PATHWAY
GO
PANTHER
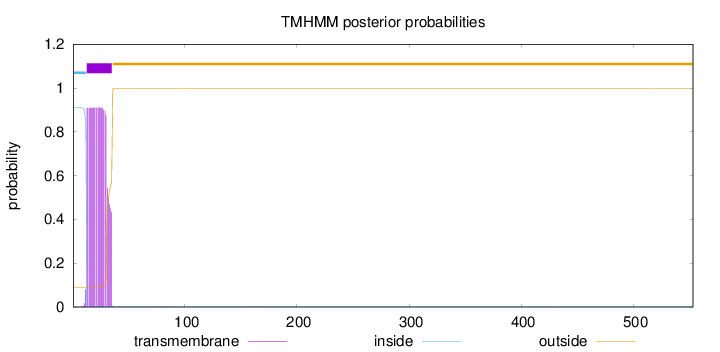
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 27,
Likelihood: 0.988804

Length:
553
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.8429999999999
Exp number, first 60 AAs:
18.83389
Total prob of N-in:
0.91045
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 553
Population Genetic Test Statistics
Pi
234.040021
Theta
180.852395
Tajima's D
0.991712
CLR
0.437151
CSRT
0.656767161641918
Interpretation
Uncertain
