Gene
KWMTBOMO13067 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000300
Annotation
PREDICTED:_eIF2B-gamma_protein_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.165 Nuclear Reliability : 1.23
Sequence
CDS
ATGCATAAAATCTTAGAATTCCAAGTTGTCGTGTTGGCCGCTGGAAAAGGTTCCCGTATGCCAGATGTCGGAGGATCAGTTTCGAAATGTCTCCTACCTGTTGGGCCGTACCCAGTACTATGGTATCCATTAAATATGCTAGAAAAAATTGGATTTCAAGATGTAATGATTGTGGTACTTGATGAAGATAAGTCTAACATTCTTAATGCACTAGAAAAATGTCCCTTGAAAATAAAATATGAGTTAATTGTAATTCCATCAGAAGAAGACTGGGGTACAGCAAATTCCTTAAAGCATGTTAGTGCTAGAATCAATACAGATTTACTAGTCATATCAGGAGACCTCATTACTAACATAAACCTCAATGATGTACTGAACCTACACAGGAAACATGATGCATGTGTCACCACCTTGTTCTTCAATAATGGTCCTGAAGAGTGGATTGAATTACCGGGACCGAAAACTAAATCCAAACCTGATCGAGACTTAGTTTGTATAGATAAAGAGACTGAGAGACTTGTTTTTCTGGCAAGTGCTAGTGATTTTGAAGAGAATGTGACAATACCAAGATTACTTGTGAAAAAATATGATGCTTTGAGCATATATAGTAGATTATTGGATGCACATGTATATGTAATGAAACACTGGATTTTAGACTATATTGTAGATTCCGAAAAATTTACCTCGATTAAAGGTGAGGTTGTTCCTTACATTGTTAAAAAGCAATTAACGAAACCTAACAACTTAGTGGAAAAGAAGGGCACCTCGGAAAAGAATGCAGAAATTAATAAAGGAATTTTTGACTATGCAATAGAGACAGGTTATGAGCGTAAGATAAGAGAAATATCAGCATACAATGATCATAAGCATGGTAACAAAGGGGTGTACTTTAATGATACTCTTAGATGTTATGCACATATTCCTAGTAAAAATACATTTGCAATTCGTGTTAATACACTATCATCATTTTATCTATCCAACAATAAGATATTATCAAAATGGCAAGATCTTACTGGATCATCTCTGTTTGAAAGATTTCACCCTAACAGTGAGGTGAAAACAAAACAGATTGATGATAATTGCACTGTTGGAGAGAAAACTATTATTAATGAAAAAACATCAGTAAAAAATTCATTTATTGGCTCAAATTGTAATATAGAAAACAAAGTCAGACTAACAAATTGTATACTCATGAACAATGTGACCATCAAAGAAAGCTGTGTTCTACAAGATTGTGTTGTCTATACCGGAGCGACAGTAGGCACCAATTGTTCAATGCAATACTGTCTTATAGGACCTCACCATTTGGTTCCGGAATCAACTAAGAGCAACCATCAACTGCATGCAGAAACTACAGATAACATGATAACGCTTGGATAA
Protein
MHKILEFQVVVLAAGKGSRMPDVGGSVSKCLLPVGPYPVLWYPLNMLEKIGFQDVMIVVLDEDKSNILNALEKCPLKIKYELIVIPSEEDWGTANSLKHVSARINTDLLVISGDLITNINLNDVLNLHRKHDACVTTLFFNNGPEEWIELPGPKTKSKPDRDLVCIDKETERLVFLASASDFEENVTIPRLLVKKYDALSIYSRLLDAHVYVMKHWILDYIVDSEKFTSIKGEVVPYIVKKQLTKPNNLVEKKGTSEKNAEINKGIFDYAIETGYERKIREISAYNDHKHGNKGVYFNDTLRCYAHIPSKNTFAIRVNTLSSFYLSNNKILSKWQDLTGSSLFERFHPNSEVKTKQIDDNCTVGEKTIINEKTSVKNSFIGSNCNIENKVRLTNCILMNNVTIKESCVLQDCVVYTGATVGTNCSMQYCLIGPHHLVPESTKSNHQLHAETTDNMITLG
Summary
Uniprot
Q0ZB79
H9ISS3
A0A2H1V500
A0A2A4IRW3
A0A2W1BJB3
A0A212FL00
+ More
S4PGV6 A0A194PXT4 A0A0L7L0U1 A0A3S2LD78 A0A194R6F4 A0A2J7RLQ5 D6X364 A0A1Y1KB30 A0A0T6B4Q4 A0A110A0M4 K7J6V8 A0A232EL02 A0A158N918 A0A1B6GVP3 U5EY76 A0A336L7M7 A0A026WT39 A0A1B6K928 A0A195BD61 F4WM19 A0A195C858 E1ZYD1 A0A154PC11 A0A2P8XSN7 A0A336MTR2 A0A195DCQ8 A0A034WEA6 A0A0A1XII4 E2B3K2 A0A3F2Z4N7 A0A0L7RAT8 A0A0V0G9L2 A0A195FW32 A0A1A9YGE4 T1PIV6 A0A1A9V2T3 A0A1I8M3S0 A0A151XA03 A0A023F973 W8AVS7 A0A1B0AAW1 A0A310SE37 A0A0A9WNQ6 A0A182JKQ4 A0A0K8TF09 A0A0M8ZXW6 A0A0C9QN18 A0A1I8PHZ5 A0A0L0C304 A0A087ZP24 E9IVG4 A0A182WK28 R4G4U7 A0A0P4VSI6 A0A1A9W9Q5 A0A182TID6 A0A182XEV9 Q7Q9J1 A0A182HNY4 A0A182YC45 A0A182P8S0 A0A182VFY9 A0A182T4G4 A0A182RKV4 A0A0K8TSE1 Q28YM8 A0A1S3JL74 A0A182GGI8 A0A182M0N2 A0A0M3QV05 B3MHH7 A0A3B0J7B1 A0A1W4WBD6 W5J742 A0A2M4AN52 Q16Q28 A0A1L8DSE4 A0A1B0G0E6 A0A2M4BNP6 A0A2M4AP68 A0A1S4FTP5 A0A2M3Z9U4 B4P7B9 B3NQB6 A0A182Q0A2 B4HS55 B4QGW1 A0A2M3ZII9 Q8MPP2 B4NMT4 A0A0P5F9R5 E9HCA4 A0A0P5KDV8 A0A182FCY3
S4PGV6 A0A194PXT4 A0A0L7L0U1 A0A3S2LD78 A0A194R6F4 A0A2J7RLQ5 D6X364 A0A1Y1KB30 A0A0T6B4Q4 A0A110A0M4 K7J6V8 A0A232EL02 A0A158N918 A0A1B6GVP3 U5EY76 A0A336L7M7 A0A026WT39 A0A1B6K928 A0A195BD61 F4WM19 A0A195C858 E1ZYD1 A0A154PC11 A0A2P8XSN7 A0A336MTR2 A0A195DCQ8 A0A034WEA6 A0A0A1XII4 E2B3K2 A0A3F2Z4N7 A0A0L7RAT8 A0A0V0G9L2 A0A195FW32 A0A1A9YGE4 T1PIV6 A0A1A9V2T3 A0A1I8M3S0 A0A151XA03 A0A023F973 W8AVS7 A0A1B0AAW1 A0A310SE37 A0A0A9WNQ6 A0A182JKQ4 A0A0K8TF09 A0A0M8ZXW6 A0A0C9QN18 A0A1I8PHZ5 A0A0L0C304 A0A087ZP24 E9IVG4 A0A182WK28 R4G4U7 A0A0P4VSI6 A0A1A9W9Q5 A0A182TID6 A0A182XEV9 Q7Q9J1 A0A182HNY4 A0A182YC45 A0A182P8S0 A0A182VFY9 A0A182T4G4 A0A182RKV4 A0A0K8TSE1 Q28YM8 A0A1S3JL74 A0A182GGI8 A0A182M0N2 A0A0M3QV05 B3MHH7 A0A3B0J7B1 A0A1W4WBD6 W5J742 A0A2M4AN52 Q16Q28 A0A1L8DSE4 A0A1B0G0E6 A0A2M4BNP6 A0A2M4AP68 A0A1S4FTP5 A0A2M3Z9U4 B4P7B9 B3NQB6 A0A182Q0A2 B4HS55 B4QGW1 A0A2M3ZII9 Q8MPP2 B4NMT4 A0A0P5F9R5 E9HCA4 A0A0P5KDV8 A0A182FCY3
Pubmed
19121390
28756777
22118469
23622113
26354079
26227816
+ More
18362917 19820115 28004739 20075255 28648823 21347285 24508170 30249741 21719571 20798317 29403074 25348373 25830018 25315136 25474469 24495485 25401762 26823975 26108605 21282665 27129103 12364791 14747013 17210077 25244985 26369729 15632085 17994087 26483478 20920257 23761445 17510324 17550304 22936249 21292972
18362917 19820115 28004739 20075255 28648823 21347285 24508170 30249741 21719571 20798317 29403074 25348373 25830018 25315136 25474469 24495485 25401762 26823975 26108605 21282665 27129103 12364791 14747013 17210077 25244985 26369729 15632085 17994087 26483478 20920257 23761445 17510324 17550304 22936249 21292972
EMBL
DQ645457
ABG54285.1
BABH01013191
ODYU01000702
SOQ35913.1
NWSH01008538
+ More
PCG62515.1 KZ150009 PZC75129.1 AGBW02007934 OWR54412.1 GAIX01003482 JAA89078.1 KQ459586 KPI97838.1 JTDY01003768 KOB69035.1 RSAL01000037 RVE51153.1 KQ460883 KPJ11446.1 NEVH01002684 PNF41772.1 KQ971372 EFA10335.1 GEZM01087386 JAV58722.1 LJIG01009855 KRT82246.1 KT768112 AMH84307.1 AAZX01001296 NNAY01003671 OXU19025.1 ADTU01001365 GECZ01003255 JAS66514.1 GANO01000538 JAB59333.1 UFQS01002342 UFQT01002342 SSX13818.1 SSX33239.1 KK107109 QOIP01000008 EZA59177.1 RLU19621.1 GEBQ01032037 JAT07940.1 KQ976519 KYM82134.1 GL888217 EGI64663.1 KQ978220 KYM96323.1 GL435204 EFN73830.1 KQ434857 KZC08808.1 PYGN01001413 PSN35021.1 UFQT01002518 SSX33616.1 KQ980989 KYN10622.1 GAKP01006844 JAC52108.1 GBXI01003093 JAD11199.1 GL445346 EFN89754.1 JXJN01005906 KQ414618 KOC67940.1 GECL01001403 JAP04721.1 KQ981264 KYN44049.1 KA648045 AFP62674.1 KQ982353 KYQ57183.1 GBBI01001254 JAC17458.1 GAMC01021669 JAB84886.1 KQ762329 OAD55945.1 GBHO01034543 GDHC01010778 JAG09061.1 JAQ07851.1 GBRD01001663 JAG64158.1 KQ435798 KOX73345.1 GBYB01004959 JAG74726.1 JRES01000975 KNC26622.1 GL766234 EFZ15436.1 ACPB03020046 GAHY01000446 JAA77064.1 GDKW01000494 JAI56101.1 AAAB01008900 EAA09498.3 APCN01002812 GDAI01000547 JAI17056.1 CM000071 EAL25937.2 JXUM01061825 KQ562170 KXJ76516.1 AXCM01001014 CP012524 ALC41543.1 CH902619 EDV37977.1 OUUW01000001 SPP75782.1 ADMH02002097 ETN59213.1 GGFK01008892 MBW42213.1 CH477765 EAT36481.1 GFDF01004763 JAV09321.1 CCAG010016559 GGFJ01005257 MBW54398.1 GGFK01009263 MBW42584.1 GGFM01004556 MBW25307.1 CM000158 EDW92064.1 CH954179 EDV55892.1 AXCN02001020 CH480816 EDW48002.1 CM000362 CM002911 EDX07224.1 KMY94012.1 GGFM01007590 MBW28341.1 AJ344148 CAC82993.1 CH964282 EDW85673.1 GDIP01207936 GDIP01150103 GDIQ01242842 GDIQ01170373 GDIQ01168567 GDIQ01166534 GDIQ01115145 GDIQ01067811 GDIQ01067810 LRGB01001569 JAJ73299.1 JAK08883.1 KZS11757.1 GL732619 EFX70645.1 GDIQ01191999 JAK59726.1
PCG62515.1 KZ150009 PZC75129.1 AGBW02007934 OWR54412.1 GAIX01003482 JAA89078.1 KQ459586 KPI97838.1 JTDY01003768 KOB69035.1 RSAL01000037 RVE51153.1 KQ460883 KPJ11446.1 NEVH01002684 PNF41772.1 KQ971372 EFA10335.1 GEZM01087386 JAV58722.1 LJIG01009855 KRT82246.1 KT768112 AMH84307.1 AAZX01001296 NNAY01003671 OXU19025.1 ADTU01001365 GECZ01003255 JAS66514.1 GANO01000538 JAB59333.1 UFQS01002342 UFQT01002342 SSX13818.1 SSX33239.1 KK107109 QOIP01000008 EZA59177.1 RLU19621.1 GEBQ01032037 JAT07940.1 KQ976519 KYM82134.1 GL888217 EGI64663.1 KQ978220 KYM96323.1 GL435204 EFN73830.1 KQ434857 KZC08808.1 PYGN01001413 PSN35021.1 UFQT01002518 SSX33616.1 KQ980989 KYN10622.1 GAKP01006844 JAC52108.1 GBXI01003093 JAD11199.1 GL445346 EFN89754.1 JXJN01005906 KQ414618 KOC67940.1 GECL01001403 JAP04721.1 KQ981264 KYN44049.1 KA648045 AFP62674.1 KQ982353 KYQ57183.1 GBBI01001254 JAC17458.1 GAMC01021669 JAB84886.1 KQ762329 OAD55945.1 GBHO01034543 GDHC01010778 JAG09061.1 JAQ07851.1 GBRD01001663 JAG64158.1 KQ435798 KOX73345.1 GBYB01004959 JAG74726.1 JRES01000975 KNC26622.1 GL766234 EFZ15436.1 ACPB03020046 GAHY01000446 JAA77064.1 GDKW01000494 JAI56101.1 AAAB01008900 EAA09498.3 APCN01002812 GDAI01000547 JAI17056.1 CM000071 EAL25937.2 JXUM01061825 KQ562170 KXJ76516.1 AXCM01001014 CP012524 ALC41543.1 CH902619 EDV37977.1 OUUW01000001 SPP75782.1 ADMH02002097 ETN59213.1 GGFK01008892 MBW42213.1 CH477765 EAT36481.1 GFDF01004763 JAV09321.1 CCAG010016559 GGFJ01005257 MBW54398.1 GGFK01009263 MBW42584.1 GGFM01004556 MBW25307.1 CM000158 EDW92064.1 CH954179 EDV55892.1 AXCN02001020 CH480816 EDW48002.1 CM000362 CM002911 EDX07224.1 KMY94012.1 GGFM01007590 MBW28341.1 AJ344148 CAC82993.1 CH964282 EDW85673.1 GDIP01207936 GDIP01150103 GDIQ01242842 GDIQ01170373 GDIQ01168567 GDIQ01166534 GDIQ01115145 GDIQ01067811 GDIQ01067810 LRGB01001569 JAJ73299.1 JAK08883.1 KZS11757.1 GL732619 EFX70645.1 GDIQ01191999 JAK59726.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000037510
UP000283053
+ More
UP000053240 UP000235965 UP000007266 UP000002358 UP000215335 UP000005205 UP000053097 UP000279307 UP000078540 UP000007755 UP000078542 UP000000311 UP000076502 UP000245037 UP000078492 UP000008237 UP000092460 UP000053825 UP000078541 UP000092443 UP000078200 UP000095301 UP000075809 UP000092445 UP000075880 UP000053105 UP000095300 UP000037069 UP000005203 UP000075920 UP000015103 UP000091820 UP000075902 UP000076407 UP000007062 UP000075840 UP000076408 UP000075885 UP000075903 UP000075901 UP000075900 UP000001819 UP000085678 UP000069940 UP000249989 UP000075883 UP000092553 UP000007801 UP000268350 UP000192221 UP000000673 UP000008820 UP000092444 UP000002282 UP000008711 UP000075886 UP000001292 UP000000304 UP000007798 UP000076858 UP000000305 UP000069272
UP000053240 UP000235965 UP000007266 UP000002358 UP000215335 UP000005205 UP000053097 UP000279307 UP000078540 UP000007755 UP000078542 UP000000311 UP000076502 UP000245037 UP000078492 UP000008237 UP000092460 UP000053825 UP000078541 UP000092443 UP000078200 UP000095301 UP000075809 UP000092445 UP000075880 UP000053105 UP000095300 UP000037069 UP000005203 UP000075920 UP000015103 UP000091820 UP000075902 UP000076407 UP000007062 UP000075840 UP000076408 UP000075885 UP000075903 UP000075901 UP000075900 UP000001819 UP000085678 UP000069940 UP000249989 UP000075883 UP000092553 UP000007801 UP000268350 UP000192221 UP000000673 UP000008820 UP000092444 UP000002282 UP000008711 UP000075886 UP000001292 UP000000304 UP000007798 UP000076858 UP000000305 UP000069272
Interpro
Gene 3D
ProteinModelPortal
Q0ZB79
H9ISS3
A0A2H1V500
A0A2A4IRW3
A0A2W1BJB3
A0A212FL00
+ More
S4PGV6 A0A194PXT4 A0A0L7L0U1 A0A3S2LD78 A0A194R6F4 A0A2J7RLQ5 D6X364 A0A1Y1KB30 A0A0T6B4Q4 A0A110A0M4 K7J6V8 A0A232EL02 A0A158N918 A0A1B6GVP3 U5EY76 A0A336L7M7 A0A026WT39 A0A1B6K928 A0A195BD61 F4WM19 A0A195C858 E1ZYD1 A0A154PC11 A0A2P8XSN7 A0A336MTR2 A0A195DCQ8 A0A034WEA6 A0A0A1XII4 E2B3K2 A0A3F2Z4N7 A0A0L7RAT8 A0A0V0G9L2 A0A195FW32 A0A1A9YGE4 T1PIV6 A0A1A9V2T3 A0A1I8M3S0 A0A151XA03 A0A023F973 W8AVS7 A0A1B0AAW1 A0A310SE37 A0A0A9WNQ6 A0A182JKQ4 A0A0K8TF09 A0A0M8ZXW6 A0A0C9QN18 A0A1I8PHZ5 A0A0L0C304 A0A087ZP24 E9IVG4 A0A182WK28 R4G4U7 A0A0P4VSI6 A0A1A9W9Q5 A0A182TID6 A0A182XEV9 Q7Q9J1 A0A182HNY4 A0A182YC45 A0A182P8S0 A0A182VFY9 A0A182T4G4 A0A182RKV4 A0A0K8TSE1 Q28YM8 A0A1S3JL74 A0A182GGI8 A0A182M0N2 A0A0M3QV05 B3MHH7 A0A3B0J7B1 A0A1W4WBD6 W5J742 A0A2M4AN52 Q16Q28 A0A1L8DSE4 A0A1B0G0E6 A0A2M4BNP6 A0A2M4AP68 A0A1S4FTP5 A0A2M3Z9U4 B4P7B9 B3NQB6 A0A182Q0A2 B4HS55 B4QGW1 A0A2M3ZII9 Q8MPP2 B4NMT4 A0A0P5F9R5 E9HCA4 A0A0P5KDV8 A0A182FCY3
S4PGV6 A0A194PXT4 A0A0L7L0U1 A0A3S2LD78 A0A194R6F4 A0A2J7RLQ5 D6X364 A0A1Y1KB30 A0A0T6B4Q4 A0A110A0M4 K7J6V8 A0A232EL02 A0A158N918 A0A1B6GVP3 U5EY76 A0A336L7M7 A0A026WT39 A0A1B6K928 A0A195BD61 F4WM19 A0A195C858 E1ZYD1 A0A154PC11 A0A2P8XSN7 A0A336MTR2 A0A195DCQ8 A0A034WEA6 A0A0A1XII4 E2B3K2 A0A3F2Z4N7 A0A0L7RAT8 A0A0V0G9L2 A0A195FW32 A0A1A9YGE4 T1PIV6 A0A1A9V2T3 A0A1I8M3S0 A0A151XA03 A0A023F973 W8AVS7 A0A1B0AAW1 A0A310SE37 A0A0A9WNQ6 A0A182JKQ4 A0A0K8TF09 A0A0M8ZXW6 A0A0C9QN18 A0A1I8PHZ5 A0A0L0C304 A0A087ZP24 E9IVG4 A0A182WK28 R4G4U7 A0A0P4VSI6 A0A1A9W9Q5 A0A182TID6 A0A182XEV9 Q7Q9J1 A0A182HNY4 A0A182YC45 A0A182P8S0 A0A182VFY9 A0A182T4G4 A0A182RKV4 A0A0K8TSE1 Q28YM8 A0A1S3JL74 A0A182GGI8 A0A182M0N2 A0A0M3QV05 B3MHH7 A0A3B0J7B1 A0A1W4WBD6 W5J742 A0A2M4AN52 Q16Q28 A0A1L8DSE4 A0A1B0G0E6 A0A2M4BNP6 A0A2M4AP68 A0A1S4FTP5 A0A2M3Z9U4 B4P7B9 B3NQB6 A0A182Q0A2 B4HS55 B4QGW1 A0A2M3ZII9 Q8MPP2 B4NMT4 A0A0P5F9R5 E9HCA4 A0A0P5KDV8 A0A182FCY3
PDB
6O9Z
E-value=3.13127e-66,
Score=640
Ontologies
KEGG
Topology
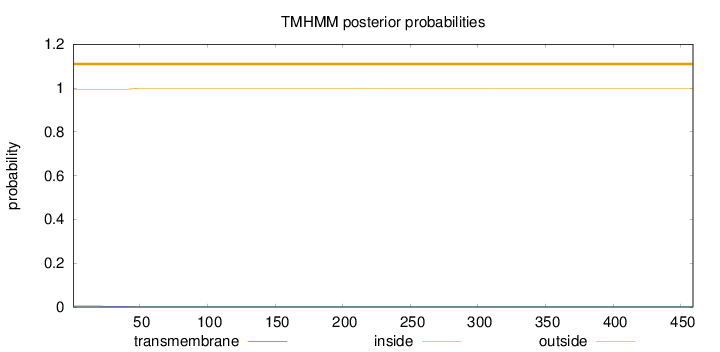
Length:
459
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0733199999999999
Exp number, first 60 AAs:
0.07123
Total prob of N-in:
0.00556
outside
1 - 459
Population Genetic Test Statistics
Pi
282.672938
Theta
182.666779
Tajima's D
1.493401
CLR
0.204316
CSRT
0.787710614469276
Interpretation
Uncertain
