Gene
KWMTBOMO13065
Pre Gene Modal
BGIBMGA000401
Annotation
PREDICTED:_uncharacterized_protein_LOC101741124_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.641
Sequence
CDS
ATGCGGTCGGCGCTTTACGCAGCATGGTGCATTTATGCGGCTTGGTTCGCGGTGTTCGCGGCGACCACTGCTGGTCGCGAGCTCCGCGTGCTACAAGCGGACGCGCGCGCCGCTTTCTACTTAGGCATCGCTGCGCCTCCCTCGGCCTCCGCCGTCGTGCGCGCGTTCAACCGATCTCTCGCGGAAGTTGCACAGACGCACCTGGCCGCCGACTACCCGCTTTACCTGCGCAACATTTCACTACTCCCCCTTTACATCGAGTTGCCAGAAGATGAAAGGTTCAATACCCGTGTTCTGGAGCGTACATGCTCAACACTGGAAGATCGACGCGTCGCCGCCTTGCTGGTCTGCGGGGGCGGATCTGCTGCAGCGGCAGTGGTGACGGCGGCAACTAGAGCTGGCGTTCCCGTACTTCGCACTACTCCTTCGCATCTGCATCTCGCCTACACTATAGCTAACGATATGCTACCTCTAGAAATAAGACTCGAAATTACTGCGCGGGAAACTTTGCAAGCACTCAGAGCAACCTTATTACATACACACTGGCATACGTTTACATTATTAGCCGAAGAAGATATACATTCTACATTATTTCTTCGAAAAGATCTTTTTCCTATTTTGTCGGCTCAGCCATTAAACCCGAAATGGCTTACGCTTCCTTCTAAATATTCACGACATATAATTTTCAGGAGACTAGCAGAAATTTCAAGGCTTACCAGAGGCATTGTTGTACTGATATGCGACATCCATTACGCTAAGCTCGTTATAGATGAAGCTAAACGCCTTAACATGCTTGATGGACATTTTTTCTGGCTTTGGATTGATGCTTCAAAAGATTTTGATGTTTTTCACAACATCAACGATAAAACTCAATTTGTTGAGGATGACATTGCTGAGTTTGAAAATATGAAAGATGGGAGTGAAAACTTTGCTAAAGACAGCTTTGTTCGCAATAAGCGTAATGATGTAGACAATAATACAGTAAATGATTTCCTAATGAATCCCACGGTGCATGAGTCTTCAGGAGATAAGCTTAGAGATCCAATTGAGAAAAGAAGTGAAAAACTGTTTACGGAGAAATTTAGAAGTACTGAAGAAAGCCATGGAATAAAAAATGCAACTTTAATTTTCAACAGTCTTCCCGTAGGACTTCTGGCTTTGCATCCACAGCCGATGAAAATAGATCGTGTATATATTCGTGCAGCAGTAAGATTAATGGTTGGAGCATTACGTCGTGTTTTGCACGCTTGCGACGCTTGGTCTGCACAAGCACAGTTCTTCAGTGACGCAACAGCATCATGCTGGGATGAACCGAGTGATGTAGCTGCAGATTTTTCATTGGAATTTGTAAGAGAGACAAGATTAGCAACAGAAGCTGTCTTATCAGGTGGTACAGAAAGGCGGGAAAGTGCACTCATGGCCAAATTTGCATTTCTAAACCTAGTTCAAGGCACAGATGGTAATAATATTTGGAGGCAAGTTGGGCAAGTAGAGGGTCGTAATGTTCGACTTCATACGATAGTTTGGCCGGGTGGAAGATTTGTTGCACATGGACATTCTGATGGAGCAAGAACAATATTTCGTATTGTCACGGCTCTTGCACCGCCTTTTGTAATGGAAGGAGAATTAGATGAAGACGGTCAATGTCTAAGAGGATTATTGTGTCATCGACCGCAAACGTCAGATCGTGATAATCTAACCCTTGCATTCAATGAATTAGAAAGGGACACTGATCCGGAGCCTGATGATTTTTTCTTTCCAACACCTAAACCAACTGTAACAAAAATGGCCACACATTGTTGTTACGGCTTAGCCATGGATCTTTTAGAAAATATTGCACAAGAACTTGAATTTGATTTCCATTTATACATTGTGGAAGACGGTGCCTACGGATCAAGACAACTTGTAAAAACTTTTCGCAGTTTTCACGAATATACTAGACCTACAGACAAATATATGACTTTGCACGATGAAAACTACAGGTCACAATATAGAAACGATTACACTATCTTACAACAGACGTCTGAGATACCGTTAATTTCCGATGACCTCGAAGATGAGGACGTAATGAAATGGAATGGGGTTGTAGGAGATTTAGTTTCTGGAGCTGCGCATATGTCCTTTGCTGCCTTAAGCGTCTCTTCAGCTAGAGCGGAAGTTATCGATTTTAGTCAACCTTATTTTTTTAGCGGTATTTCCGTATTGGCAGCTCCAAATCAAAGACCAGATATACCTTTACTAGCTTTTCTTTTGCCCTTTTCGCCGGAGTTGTGGATTGCAATATTTACATCGTTAAACGTGACTGCGATTGCCGTTGCAATATATGAATGGTTGTCACCATTTGGACTGAATCCGTGGGGTCGTCAGAGATCAAAGAATTTTAGTATTTCCAGTGCTCTGTGGGTGATGTGGGGGCTCCTGTGTGGGCATTTGGTAGCATTTAAAGCACCCAAAAGTTGGCCTAATAAGTTTCTTATCAACGTCTGGGGTGGATTTTCTGTAATATTTGTTGCAAGTTATACTGCAAACATAGCTGCACTTATAGCTGGATTATTTTTTCATAACGCTGTCGATGATTTTCAAGGAGGTGATAATTGGCTTTCGCTTAAAGTGGGGACCGCAAGATCATCCGTAGCTGAATATTATGTTCAAAGAAATAATCCGCATTTGGCCCAGCAGATGCGTCGCTATGCCCTCCAGGATATCGAAGAGGGTATTCAACGGCTTAGAAACGGAACACTAGATCTCTTGATATCAGACACTCCTGTTTTGGATTACTATAGAGCAACCGATCATGGATGTAAGCTTCAACGTGTGGGCGGTCGTACTCTGGCAGAAGATACCTATGCTATTGGCATGACAAAAGGATTTCCATTGAAGGACAGTATATCTTCAGTTATAGCTAAGTATTCGTCGAATGGATATATGGACATATTAACTGAAAAATGGTACGGCGGTCTACCTTGTTTTAAACTCAGCCCAGATTATGGCATTCAACCGAAACCTTTAGGTGTGGCGGCTGTGGCTGGTGTTTTTATTCTTCTTGGAGTAGGAATGATCGTTGGAATTATCATTTTGATTTTCGAACATCTGTTTTATAAATACACTTTACCTATATTAAGGCATCAACCGAAAGGAACAATCTGGAGGAGCAGGAACATTATGTTTTTCAGTCAGAAACTATACCGATTTATCAATTGTGTTGAATTGGTATCGCCGCACCATGCCGCCCGAGAATTAGTCAATACAATACGCCAAGGTCATTTCACTTCTCTGTTCCAAAAGAGCGTTAAAAGGAAGGAACATGAACAGCGCAGGCGTCGTAAGAGTAAAGCGCAATTGTACGAAATGATTCAAGCGATTAGAAGAGTACAACAACGGGATCACTCTTTGGGGTCTATTAAAGAACAAGAGCCAGTTGATACATCTGAGACAACAGAAGAATTAACTGAATCAAAGTTTTTATCCCCGTCACCCGACACATCGCACCGCTCACCAAGACAAGGTCGTTCACCTCGACAGCTTCGTTCGCCTAAAGGTCGTCGAAAAAGATGCAGTCTCGCAGGATTAAACGTTAGGAGATTTAGTACCGATTCAGTGTTGGGTTCAGATTCTGTTTCCAATATTTATGAGAGAACTTGCCACAACATAGGCAGACGACTTAGTCGAGATGTCAGCTGCTTAACAAATTCCCCTCCAGATTTAAACACCCGATTGCGAACACCGTCTCCGATGATAAGACGGACCGAAGCTTCTTCAACCCGTTCTTACCAAGACGTATCCTTAAGATCCGAAAATTATGTTAGCACAGATGCTCCAACGTCTCGAGCGAGTATAGATATTTTAATTTCTGATGAACATGATATACCTCCTGCGCCACCATATCCTAGAGTTTCTCCGGCGGGTGCTCGATCCGAACTTTCCTTACTATCTGAAGAAGAGTTGATTAGACTTTGGCGAAGTTCAGAACGCGAAGTCAGAGAGGCTTTATTAGCAGCACTACAAGAAAGACGGGCGAACTTAGATCCCAAAGAAGATCCGGGATGA
Protein
MRSALYAAWCIYAAWFAVFAATTAGRELRVLQADARAAFYLGIAAPPSASAVVRAFNRSLAEVAQTHLAADYPLYLRNISLLPLYIELPEDERFNTRVLERTCSTLEDRRVAALLVCGGGSAAAAVVTAATRAGVPVLRTTPSHLHLAYTIANDMLPLEIRLEITARETLQALRATLLHTHWHTFTLLAEEDIHSTLFLRKDLFPILSAQPLNPKWLTLPSKYSRHIIFRRLAEISRLTRGIVVLICDIHYAKLVIDEAKRLNMLDGHFFWLWIDASKDFDVFHNINDKTQFVEDDIAEFENMKDGSENFAKDSFVRNKRNDVDNNTVNDFLMNPTVHESSGDKLRDPIEKRSEKLFTEKFRSTEESHGIKNATLIFNSLPVGLLALHPQPMKIDRVYIRAAVRLMVGALRRVLHACDAWSAQAQFFSDATASCWDEPSDVAADFSLEFVRETRLATEAVLSGGTERRESALMAKFAFLNLVQGTDGNNIWRQVGQVEGRNVRLHTIVWPGGRFVAHGHSDGARTIFRIVTALAPPFVMEGELDEDGQCLRGLLCHRPQTSDRDNLTLAFNELERDTDPEPDDFFFPTPKPTVTKMATHCCYGLAMDLLENIAQELEFDFHLYIVEDGAYGSRQLVKTFRSFHEYTRPTDKYMTLHDENYRSQYRNDYTILQQTSEIPLISDDLEDEDVMKWNGVVGDLVSGAAHMSFAALSVSSARAEVIDFSQPYFFSGISVLAAPNQRPDIPLLAFLLPFSPELWIAIFTSLNVTAIAVAIYEWLSPFGLNPWGRQRSKNFSISSALWVMWGLLCGHLVAFKAPKSWPNKFLINVWGGFSVIFVASYTANIAALIAGLFFHNAVDDFQGGDNWLSLKVGTARSSVAEYYVQRNNPHLAQQMRRYALQDIEEGIQRLRNGTLDLLISDTPVLDYYRATDHGCKLQRVGGRTLAEDTYAIGMTKGFPLKDSISSVIAKYSSNGYMDILTEKWYGGLPCFKLSPDYGIQPKPLGVAAVAGVFILLGVGMIVGIIILIFEHLFYKYTLPILRHQPKGTIWRSRNIMFFSQKLYRFINCVELVSPHHAARELVNTIRQGHFTSLFQKSVKRKEHEQRRRRKSKAQLYEMIQAIRRVQQRDHSLGSIKEQEPVDTSETTEELTESKFLSPSPDTSHRSPRQGRSPRQLRSPKGRRKRCSLAGLNVRRFSTDSVLGSDSVSNIYERTCHNIGRRLSRDVSCLTNSPPDLNTRLRTPSPMIRRTEASSTRSYQDVSLRSENYVSTDAPTSRASIDILISDEHDIPPAPPYPRVSPAGARSELSLLSEEELIRLWRSSEREVREALLAALQERRANLDPKEDPG
Summary
Similarity
Belongs to the glutamate-gated ion channel (TC 1.A.10.1) family.
Uniprot
Pubmed
EMBL
KQ459586
KPI97834.1
KZ149941
PZC76914.1
ODYU01003983
SOQ43403.1
+ More
RSAL01000037 RVE51149.1 KQ460883 KPJ11449.1 AGBW02011767 OWR46167.1 KQ971371 EFA09473.2 DS235019 EEB10559.1 PYGN01000144 PSN52890.1 QOIP01000004 RLU24085.1 KQ982481 KYQ55990.1 GL888285 EGI63214.1 KQ981522 KYN40620.1 ADTU01026589 KQ976504 KYM82861.1 KK107024 EZA62306.1 KQ980368 KYN16338.1 KQ977771 KYN00038.1 GL765283 EFZ16607.1 KZ288266 PBC30154.1 KQ434868 KZC09190.1 GL452067 EFN77985.1
RSAL01000037 RVE51149.1 KQ460883 KPJ11449.1 AGBW02011767 OWR46167.1 KQ971371 EFA09473.2 DS235019 EEB10559.1 PYGN01000144 PSN52890.1 QOIP01000004 RLU24085.1 KQ982481 KYQ55990.1 GL888285 EGI63214.1 KQ981522 KYN40620.1 ADTU01026589 KQ976504 KYM82861.1 KK107024 EZA62306.1 KQ980368 KYN16338.1 KQ977771 KYN00038.1 GL765283 EFZ16607.1 KZ288266 PBC30154.1 KQ434868 KZC09190.1 GL452067 EFN77985.1
Proteomes
Interpro
ProteinModelPortal
PDB
6MMX
E-value=1.50347e-46,
Score=475
Ontologies
KEGG
GO
Topology
Subcellular location
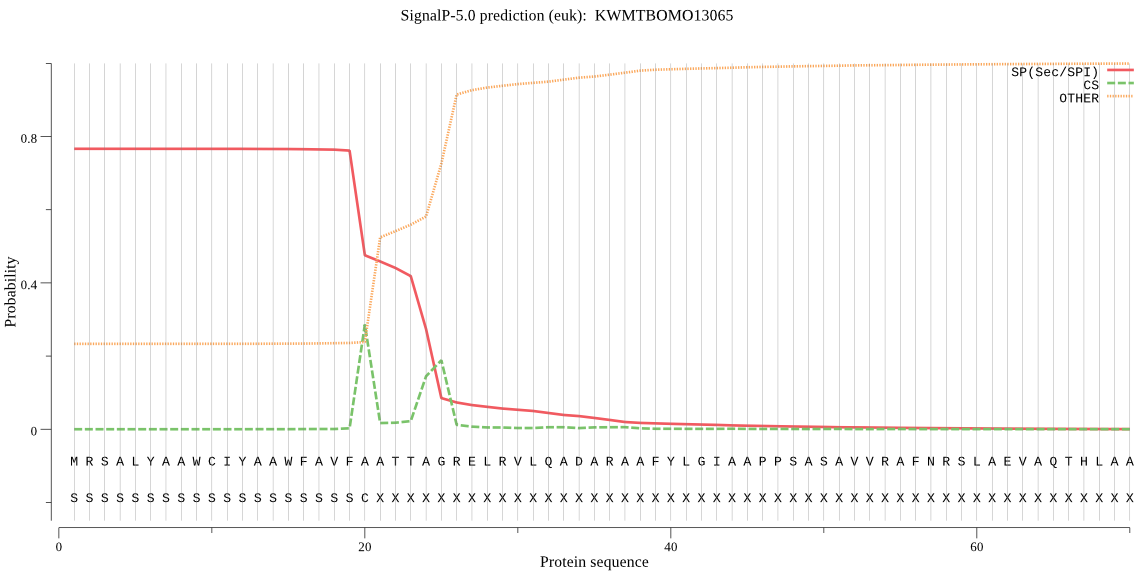
SignalP
Position: 1 - 20,
Likelihood: 0.765280
Length:
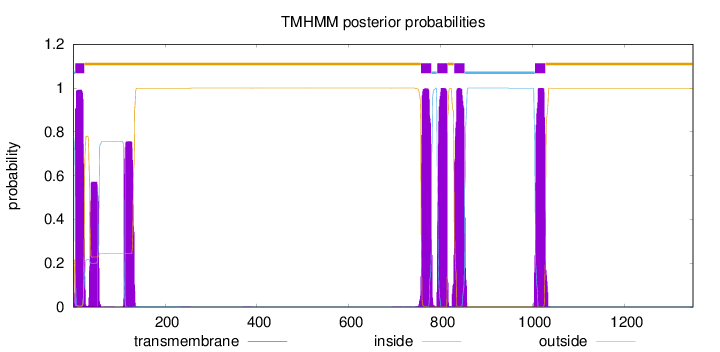
1348
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
137.77125
Exp number, first 60 AAs:
31.43918
Total prob of N-in:
0.78363
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 24
outside
25 - 756
TMhelix
757 - 779
inside
780 - 791
TMhelix
792 - 814
outside
815 - 828
TMhelix
829 - 851
inside
852 - 1004
TMhelix
1005 - 1027
outside
1028 - 1348
Population Genetic Test Statistics
Pi
235.844501
Theta
194.672597
Tajima's D
0.471467
CLR
0.448231
CSRT
0.508724563771811
Interpretation
Uncertain
