Gene
KWMTBOMO13064 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000402
Annotation
putative_multiple_inositol_polyphosphate_phosphatase_[Danaus_plexippus]
Location in the cell
Extracellular Reliability : 1.419
Sequence
CDS
ATGAAATTTCTATTCGTGACCGTGTTTCTAAGCGTTTATGCCAGATCTGTGATTTCGTTGTTCTGTTATTGGAATACCGGCTGTCCATATAATTATTTTTCTAGCAGAACTCCATATAATGCAGCCAGAGGGGATATAAGAGACTCAGTTATCAAACTTACCGATTGTGAACCAATAAGTATTTGGGGTCTTGTAAGACATGGTAAAAGGAATCCTGGTGCAGAATTAGCATTGACTATGAAGAATGCTATTGTAATCAGAGAATACGTTGTATCTAGTTATGAGAATGGAAACAGCTCATTATGCGCGCAAGATATCGAAAATTTGAGAGAATTGGGCGCTGATTATGGCATGTTCGAAAATGCATATCAACTCTCTGAAGAAGGTTATCAAGAGATGATGGATATTGGTAAAAGATTTAAACAAGCTTTTCCTAAATTATTGAACAAACTGGAAAGTCAGAGCTACACTTTTAGACCGGCTTTTGGCAAGTGGATGCAAAAAAGTGCTGAAGGATTCGTAAATGGACTCGCTAATGGTAACTTGGACATTGAAAAGGCTACTACTGATTTTGACATTATGGATCCTTACACAACATGTGGCAAATATCAAAGAGACGTAAAAAAGAATCCTGAGATATACCTCGAATCAAATAAATATTTGGAAACAACGGAATTTTTGGCTACAAAAGATAGAATTCAAAGGAGATTGGGAATCGATTATCCATTAACAAATGAAAACATATCGGCTTTGTATGACCTCTGTCGATACACTTGGTCAAGCAAAGACAAAATGAGCCCATGGTGCGCATTATTTACGACTGAGGATCTCAAGGTTTTAGAATACGCAGGAGATCTGAAACACTATTACAGAAACGGTTATGGAAACTCAATCAACGCTCATTTAGGACAGATTCCATTGAGTGATTTATTCAAAAGCTTTCAATTAGCTAAAGATGGAAAAGGAAAAAAGATAATAGCTTATTTCACTCATGCAACAATGATGGATATGTTGTACACGGCCTTAAATTTGTTTAAAGACGATGTTGAACTAACTGGTTCACTTAGAAACCCTGACAGAAAATGGAGAACTAGTAAATTGTCCATCTTCGGAGCAAATATGTTTGCAGTACTCAGCAGGTGTAATAGAGAAAATAAAACGGACTACAACGTTGTTTTTTACTTAAACGAAGAGCCACTGAAACCAATTTGTGAACAAGGAGTGTGCACATGGGAGGAATTCGAAAACAAGTTCAAAACAATGAACTCCAACACGGATATGTGTCAATTCAAAAGTCTACCATACATTTAA
Protein
MKFLFVTVFLSVYARSVISLFCYWNTGCPYNYFSSRTPYNAARGDIRDSVIKLTDCEPISIWGLVRHGKRNPGAELALTMKNAIVIREYVVSSYENGNSSLCAQDIENLRELGADYGMFENAYQLSEEGYQEMMDIGKRFKQAFPKLLNKLESQSYTFRPAFGKWMQKSAEGFVNGLANGNLDIEKATTDFDIMDPYTTCGKYQRDVKKNPEIYLESNKYLETTEFLATKDRIQRRLGIDYPLTNENISALYDLCRYTWSSKDKMSPWCALFTTEDLKVLEYAGDLKHYYRNGYGNSINAHLGQIPLSDLFKSFQLAKDGKGKKIIAYFTHATMMDMLYTALNLFKDDVELTGSLRNPDRKWRTSKLSIFGANMFAVLSRCNRENKTDYNVVFYLNEEPLKPICEQGVCTWEEFENKFKTMNSNTDMCQFKSLPYI
Summary
Similarity
Belongs to the histidine acid phosphatase family.
Uniprot
H9IT23
A0A2W1BTF1
A0A2H1VSH2
I4DNN5
A0A0L7LGE4
A0A3S2LPK0
+ More
A0A194R6G0 A0A194PXT0 A0A2H1V6V9 A0A2W1BW81 A0A2W1BZM1 A0A212EXE4 A0A2H1VP81 A0A2W1BJ61 A0A1E1W4B6 A0A194Q1Z1 A0A194RFH5 A0A3S2PAE8 A0A212ERM3 A0A0L7KTC5 A0A212EKN7 A0A212FEA4 A0A0C9RUR7 A0A067R6V0 A0A2J7RDL5 A0A2J7RDL3 A0A1B6IKU5 D6WUK9 A0A1B6EIW9 A0A0L7QR89 E9G9W2 A0A2J7RDK1 A0A2J7RDJ9 A0A067R8V8 A0A2J7RDK4 K7J1W2 A0A0P6ITU3 A0A0P4YQH5 A0A2P8XRY2 F4WHR4 A0A088AJX1 A0A2J7RDK2 E2B471 A0A158NM28 A0A0P6BS44 K7J1W3 A0A1B6DXK1 A0A1B6DSF1 A0A232FCC4 E1ZUZ6 A0A2J7RDK5 A0A195CX09 A0A0N8D4B2 E9IIG1 U5EX79 A0A0P5U8D8 A0A0P4ZC10 A0A0P4YM59 A0A1B6MC10 A0A2J7RDK9 A0A195B4H3 A0A164X172 A0A1B6KA08 A0A1B6JPG7 A0A310SND8 A0A151IXB5 A0A0P5BKZ3 A0A151JXQ6 A0A0N8DI12 T1J676 A0A0P6IXT1 A0A1B6EC57 A0A154P0U4 N6UIY3 U4UGG7 A0A067RID8 A0A0P4Y6F2 A0A151XHW5 A0A026WEW2 A0A026WG54 A0A1B6GW26 A0A1B6F1J4 A0A1B6GAP1 A0A1B6GTD8 A0A194PK41 A0A0P6HKD6 A0A2H1VIP3 A0A0L7LPL9 A0A0P5KKG6 A0A023ESJ6 E9G9W3 E0VT88 A0A1B6GP03 A0A0P5KDR1 A0A0P5N917 A0A0P5KZ62 A0A023EVE1 A0A182H850 A0A0P5PHK9
A0A194R6G0 A0A194PXT0 A0A2H1V6V9 A0A2W1BW81 A0A2W1BZM1 A0A212EXE4 A0A2H1VP81 A0A2W1BJ61 A0A1E1W4B6 A0A194Q1Z1 A0A194RFH5 A0A3S2PAE8 A0A212ERM3 A0A0L7KTC5 A0A212EKN7 A0A212FEA4 A0A0C9RUR7 A0A067R6V0 A0A2J7RDL5 A0A2J7RDL3 A0A1B6IKU5 D6WUK9 A0A1B6EIW9 A0A0L7QR89 E9G9W2 A0A2J7RDK1 A0A2J7RDJ9 A0A067R8V8 A0A2J7RDK4 K7J1W2 A0A0P6ITU3 A0A0P4YQH5 A0A2P8XRY2 F4WHR4 A0A088AJX1 A0A2J7RDK2 E2B471 A0A158NM28 A0A0P6BS44 K7J1W3 A0A1B6DXK1 A0A1B6DSF1 A0A232FCC4 E1ZUZ6 A0A2J7RDK5 A0A195CX09 A0A0N8D4B2 E9IIG1 U5EX79 A0A0P5U8D8 A0A0P4ZC10 A0A0P4YM59 A0A1B6MC10 A0A2J7RDK9 A0A195B4H3 A0A164X172 A0A1B6KA08 A0A1B6JPG7 A0A310SND8 A0A151IXB5 A0A0P5BKZ3 A0A151JXQ6 A0A0N8DI12 T1J676 A0A0P6IXT1 A0A1B6EC57 A0A154P0U4 N6UIY3 U4UGG7 A0A067RID8 A0A0P4Y6F2 A0A151XHW5 A0A026WEW2 A0A026WG54 A0A1B6GW26 A0A1B6F1J4 A0A1B6GAP1 A0A1B6GTD8 A0A194PK41 A0A0P6HKD6 A0A2H1VIP3 A0A0L7LPL9 A0A0P5KKG6 A0A023ESJ6 E9G9W3 E0VT88 A0A1B6GP03 A0A0P5KDR1 A0A0P5N917 A0A0P5KZ62 A0A023EVE1 A0A182H850 A0A0P5PHK9
Pubmed
EMBL
BABH01013207
KZ149941
KZ149892
PZC76915.1
PZC79045.1
ODYU01003983
+ More
SOQ43402.1 AK403047 BAM19525.1 JTDY01001212 KOB74522.1 RSAL01000037 RVE51148.1 KQ460883 KPJ11451.1 KQ459586 KPI97833.1 ODYU01000744 SOQ35994.1 PZC76916.1 PZC79044.1 AGBW02011767 OWR46166.1 ODYU01003636 SOQ42641.1 KZ150021 PZC74898.1 GDQN01009275 JAT81779.1 KQ459580 KPI99348.1 KQ460211 KPJ16573.1 RSAL01000147 RVE45895.1 AGBW02012983 OWR44137.1 JTDY01005875 KOB66528.1 AGBW02014223 OWR42062.1 AGBW02008959 OWR52086.1 GBYB01006096 GBYB01012605 JAG75863.1 JAG82372.1 KK852663 KDR19047.1 NEVH01005288 PNF38918.1 PNF38919.1 GECU01020168 JAS87538.1 KQ971357 EFA08487.2 GECZ01031900 JAS37869.1 KQ414784 KOC61016.1 GL732536 EFX83810.1 PNF38916.1 PNF38917.1 KDR19043.1 PNF38911.1 GDIQ01017791 JAN76946.1 GDIP01224285 GDIP01124790 JAI99116.1 PYGN01001450 PSN34766.1 GL888167 EGI66271.1 PNF38913.1 GL445515 EFN89488.1 ADTU01020207 ADTU01020208 GDIP01010892 JAM92823.1 GEDC01006884 JAS30414.1 GEDC01008715 JAS28583.1 NNAY01000444 OXU28335.1 GL434335 EFN74994.1 PNF38908.1 KQ977171 KYN05185.1 GDIP01070151 JAM33564.1 GL763491 EFZ19634.1 GANO01002665 JAB57206.1 GDIP01118197 JAL85517.1 GDIP01216140 GDIP01092605 GDIP01043270 JAJ07262.1 GDIP01226840 JAI96561.1 GEBQ01006511 JAT33466.1 PNF38912.1 KQ976604 KYM79398.1 LRGB01001036 KZS13773.1 GEBQ01031695 JAT08282.1 GECU01027339 GECU01006669 JAS80367.1 JAT01038.1 KQ759847 OAD62582.1 KQ980813 KYN12703.1 GDIP01198048 JAJ25354.1 KQ981561 KYN39831.1 GDIP01031751 JAM71964.1 JH431872 GDIQ01026114 JAN68623.1 GEDC01001805 JAS35493.1 KQ434796 KZC05546.1 APGK01018215 KB740071 ENN81710.1 KB632286 ERL91433.1 KDR19044.1 GDIP01232899 JAI90502.1 KQ982109 KYQ59996.1 KK107238 EZA54625.1 EZA54626.1 GECZ01003135 JAS66634.1 GECZ01025679 JAS44090.1 GECZ01010277 JAS59492.1 GECZ01015696 GECZ01004059 JAS54073.1 JAS65710.1 KQ459603 KPI93094.1 GDIQ01031907 JAN62830.1 ODYU01002780 SOQ40697.1 JTDY01000396 KOB77377.1 GDIQ01183946 JAK67779.1 GAPW01001348 JAC12250.1 EFX83809.1 DS235760 EEB16594.1 GECZ01005686 JAS64083.1 GDIQ01185742 JAK65983.1 GDIQ01145463 JAL06263.1 GDIQ01177248 JAK74477.1 GAPW01001159 JAC12439.1 JXUM01117635 JXUM01117636 GDIQ01138422 JAL13304.1
SOQ43402.1 AK403047 BAM19525.1 JTDY01001212 KOB74522.1 RSAL01000037 RVE51148.1 KQ460883 KPJ11451.1 KQ459586 KPI97833.1 ODYU01000744 SOQ35994.1 PZC76916.1 PZC79044.1 AGBW02011767 OWR46166.1 ODYU01003636 SOQ42641.1 KZ150021 PZC74898.1 GDQN01009275 JAT81779.1 KQ459580 KPI99348.1 KQ460211 KPJ16573.1 RSAL01000147 RVE45895.1 AGBW02012983 OWR44137.1 JTDY01005875 KOB66528.1 AGBW02014223 OWR42062.1 AGBW02008959 OWR52086.1 GBYB01006096 GBYB01012605 JAG75863.1 JAG82372.1 KK852663 KDR19047.1 NEVH01005288 PNF38918.1 PNF38919.1 GECU01020168 JAS87538.1 KQ971357 EFA08487.2 GECZ01031900 JAS37869.1 KQ414784 KOC61016.1 GL732536 EFX83810.1 PNF38916.1 PNF38917.1 KDR19043.1 PNF38911.1 GDIQ01017791 JAN76946.1 GDIP01224285 GDIP01124790 JAI99116.1 PYGN01001450 PSN34766.1 GL888167 EGI66271.1 PNF38913.1 GL445515 EFN89488.1 ADTU01020207 ADTU01020208 GDIP01010892 JAM92823.1 GEDC01006884 JAS30414.1 GEDC01008715 JAS28583.1 NNAY01000444 OXU28335.1 GL434335 EFN74994.1 PNF38908.1 KQ977171 KYN05185.1 GDIP01070151 JAM33564.1 GL763491 EFZ19634.1 GANO01002665 JAB57206.1 GDIP01118197 JAL85517.1 GDIP01216140 GDIP01092605 GDIP01043270 JAJ07262.1 GDIP01226840 JAI96561.1 GEBQ01006511 JAT33466.1 PNF38912.1 KQ976604 KYM79398.1 LRGB01001036 KZS13773.1 GEBQ01031695 JAT08282.1 GECU01027339 GECU01006669 JAS80367.1 JAT01038.1 KQ759847 OAD62582.1 KQ980813 KYN12703.1 GDIP01198048 JAJ25354.1 KQ981561 KYN39831.1 GDIP01031751 JAM71964.1 JH431872 GDIQ01026114 JAN68623.1 GEDC01001805 JAS35493.1 KQ434796 KZC05546.1 APGK01018215 KB740071 ENN81710.1 KB632286 ERL91433.1 KDR19044.1 GDIP01232899 JAI90502.1 KQ982109 KYQ59996.1 KK107238 EZA54625.1 EZA54626.1 GECZ01003135 JAS66634.1 GECZ01025679 JAS44090.1 GECZ01010277 JAS59492.1 GECZ01015696 GECZ01004059 JAS54073.1 JAS65710.1 KQ459603 KPI93094.1 GDIQ01031907 JAN62830.1 ODYU01002780 SOQ40697.1 JTDY01000396 KOB77377.1 GDIQ01183946 JAK67779.1 GAPW01001348 JAC12250.1 EFX83809.1 DS235760 EEB16594.1 GECZ01005686 JAS64083.1 GDIQ01185742 JAK65983.1 GDIQ01145463 JAL06263.1 GDIQ01177248 JAK74477.1 GAPW01001159 JAC12439.1 JXUM01117635 JXUM01117636 GDIQ01138422 JAL13304.1
Proteomes
UP000005204
UP000037510
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000027135 UP000235965 UP000007266 UP000053825 UP000000305 UP000002358 UP000245037 UP000007755 UP000005203 UP000008237 UP000005205 UP000215335 UP000000311 UP000078542 UP000078540 UP000076858 UP000078492 UP000078541 UP000076502 UP000019118 UP000030742 UP000075809 UP000053097 UP000009046 UP000069940
UP000027135 UP000235965 UP000007266 UP000053825 UP000000305 UP000002358 UP000245037 UP000007755 UP000005203 UP000008237 UP000005205 UP000215335 UP000000311 UP000078542 UP000078540 UP000076858 UP000078492 UP000078541 UP000076502 UP000019118 UP000030742 UP000075809 UP000053097 UP000009046 UP000069940
PRIDE
Pfam
Interpro
IPR000560
His_Pase_clade-2
+ More
IPR029033 His_PPase_superfam
IPR016274 Histidine_acid_Pase_euk
IPR033379 Acid_Pase_AS
IPR037278 ARFGAP/RecO
IPR001164 ArfGAP_dom
IPR038508 ArfGAP_dom_sf
IPR015915 Kelch-typ_b-propeller
IPR018379 BEN_domain
IPR011333 SKP1/BTB/POZ_sf
IPR000210 BTB/POZ_dom
IPR006652 Kelch_1
IPR011705 BACK
IPR005334 Tctex-1-like
IPR038586 Tctex-1-like_sf
IPR029033 His_PPase_superfam
IPR016274 Histidine_acid_Pase_euk
IPR033379 Acid_Pase_AS
IPR037278 ARFGAP/RecO
IPR001164 ArfGAP_dom
IPR038508 ArfGAP_dom_sf
IPR015915 Kelch-typ_b-propeller
IPR018379 BEN_domain
IPR011333 SKP1/BTB/POZ_sf
IPR000210 BTB/POZ_dom
IPR006652 Kelch_1
IPR011705 BACK
IPR005334 Tctex-1-like
IPR038586 Tctex-1-like_sf
ProteinModelPortal
H9IT23
A0A2W1BTF1
A0A2H1VSH2
I4DNN5
A0A0L7LGE4
A0A3S2LPK0
+ More
A0A194R6G0 A0A194PXT0 A0A2H1V6V9 A0A2W1BW81 A0A2W1BZM1 A0A212EXE4 A0A2H1VP81 A0A2W1BJ61 A0A1E1W4B6 A0A194Q1Z1 A0A194RFH5 A0A3S2PAE8 A0A212ERM3 A0A0L7KTC5 A0A212EKN7 A0A212FEA4 A0A0C9RUR7 A0A067R6V0 A0A2J7RDL5 A0A2J7RDL3 A0A1B6IKU5 D6WUK9 A0A1B6EIW9 A0A0L7QR89 E9G9W2 A0A2J7RDK1 A0A2J7RDJ9 A0A067R8V8 A0A2J7RDK4 K7J1W2 A0A0P6ITU3 A0A0P4YQH5 A0A2P8XRY2 F4WHR4 A0A088AJX1 A0A2J7RDK2 E2B471 A0A158NM28 A0A0P6BS44 K7J1W3 A0A1B6DXK1 A0A1B6DSF1 A0A232FCC4 E1ZUZ6 A0A2J7RDK5 A0A195CX09 A0A0N8D4B2 E9IIG1 U5EX79 A0A0P5U8D8 A0A0P4ZC10 A0A0P4YM59 A0A1B6MC10 A0A2J7RDK9 A0A195B4H3 A0A164X172 A0A1B6KA08 A0A1B6JPG7 A0A310SND8 A0A151IXB5 A0A0P5BKZ3 A0A151JXQ6 A0A0N8DI12 T1J676 A0A0P6IXT1 A0A1B6EC57 A0A154P0U4 N6UIY3 U4UGG7 A0A067RID8 A0A0P4Y6F2 A0A151XHW5 A0A026WEW2 A0A026WG54 A0A1B6GW26 A0A1B6F1J4 A0A1B6GAP1 A0A1B6GTD8 A0A194PK41 A0A0P6HKD6 A0A2H1VIP3 A0A0L7LPL9 A0A0P5KKG6 A0A023ESJ6 E9G9W3 E0VT88 A0A1B6GP03 A0A0P5KDR1 A0A0P5N917 A0A0P5KZ62 A0A023EVE1 A0A182H850 A0A0P5PHK9
A0A194R6G0 A0A194PXT0 A0A2H1V6V9 A0A2W1BW81 A0A2W1BZM1 A0A212EXE4 A0A2H1VP81 A0A2W1BJ61 A0A1E1W4B6 A0A194Q1Z1 A0A194RFH5 A0A3S2PAE8 A0A212ERM3 A0A0L7KTC5 A0A212EKN7 A0A212FEA4 A0A0C9RUR7 A0A067R6V0 A0A2J7RDL5 A0A2J7RDL3 A0A1B6IKU5 D6WUK9 A0A1B6EIW9 A0A0L7QR89 E9G9W2 A0A2J7RDK1 A0A2J7RDJ9 A0A067R8V8 A0A2J7RDK4 K7J1W2 A0A0P6ITU3 A0A0P4YQH5 A0A2P8XRY2 F4WHR4 A0A088AJX1 A0A2J7RDK2 E2B471 A0A158NM28 A0A0P6BS44 K7J1W3 A0A1B6DXK1 A0A1B6DSF1 A0A232FCC4 E1ZUZ6 A0A2J7RDK5 A0A195CX09 A0A0N8D4B2 E9IIG1 U5EX79 A0A0P5U8D8 A0A0P4ZC10 A0A0P4YM59 A0A1B6MC10 A0A2J7RDK9 A0A195B4H3 A0A164X172 A0A1B6KA08 A0A1B6JPG7 A0A310SND8 A0A151IXB5 A0A0P5BKZ3 A0A151JXQ6 A0A0N8DI12 T1J676 A0A0P6IXT1 A0A1B6EC57 A0A154P0U4 N6UIY3 U4UGG7 A0A067RID8 A0A0P4Y6F2 A0A151XHW5 A0A026WEW2 A0A026WG54 A0A1B6GW26 A0A1B6F1J4 A0A1B6GAP1 A0A1B6GTD8 A0A194PK41 A0A0P6HKD6 A0A2H1VIP3 A0A0L7LPL9 A0A0P5KKG6 A0A023ESJ6 E9G9W3 E0VT88 A0A1B6GP03 A0A0P5KDR1 A0A0P5N917 A0A0P5KZ62 A0A023EVE1 A0A182H850 A0A0P5PHK9
PDB
2GFI
E-value=1.05921e-06,
Score=126
Ontologies
PATHWAY
GO
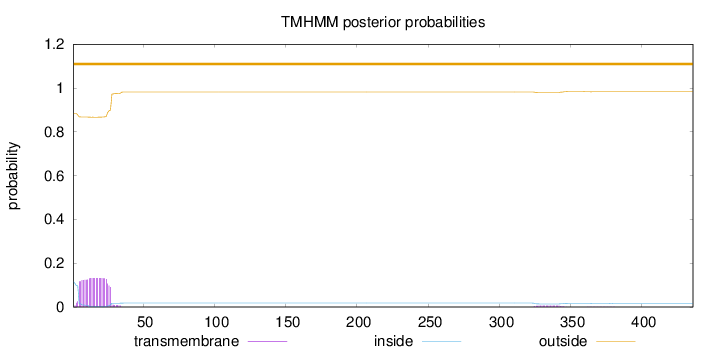
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.990027
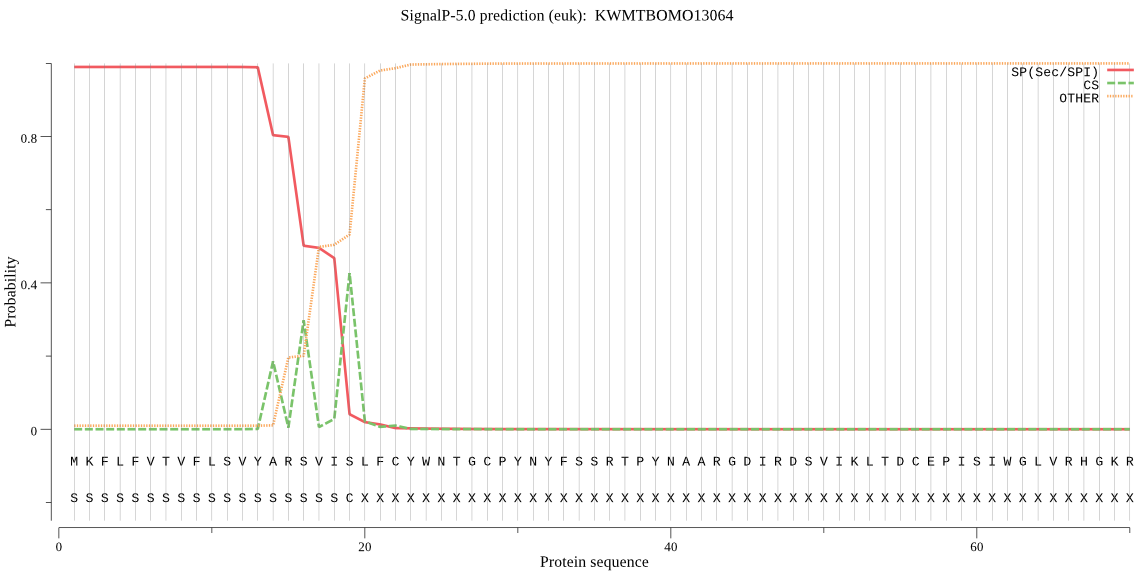
Length:
436
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.1017
Exp number, first 60 AAs:
2.96862
Total prob of N-in:
0.11580
outside
1 - 436
Population Genetic Test Statistics
Pi
229.243877
Theta
182.609281
Tajima's D
-1.716898
CLR
10.262716
CSRT
0.0381480925953702
Interpretation
Uncertain
