Gene
KWMTBOMO13062
Pre Gene Modal
BGIBMGA000295
Annotation
PREDICTED:_ras-related_GTP-binding_protein_Rab3_isoform_X2_[Bombyx_mori]
Full name
Ras-related protein Rab-3
Location in the cell
Cytoplasmic Reliability : 1.628
Sequence
CDS
ATGACTGGCGAAGCAAAATGGCAGAAAGATGCTGCGGATCAGAACTTCGATTACATGTTCAAACTGTTGATCATAGGAAACAGCTCAGTAGGAAAAACTAGTTTCCTCTTCCGATATGCAGACGATTCCTTCACTTCAGCCTTTGTGTCTACCGTTGGTATCGACTTCAAAGTGAAAACGGTTTTCCGGCACGATAAGCGCGTCAAATTGCAAATATGGGATACTGCAGGTCAGGAACGCTACCGTACCATCACGACTGCCTATTATCGAGGTGCTATGGGCTTCATTCTGATGTATGATATTACTAACGAAGAAAGTTTCAACAGCGTACAGGACTGGGTTACCCAGATCAAGACATATTCGTGGGATAATGCTCAAGTTATTCTCGTCGGAAACAAATGTGACATGGAAGAAGAACGTGTAGTTAGCGCGGAGCGTGGACGACAGTTGGCTGATCAACTGGGTGTCGAGTTTTATGAAACAAGCGCAAAAGAAAATATAAACGTCAAGGCTGTATTCGAGCGACTAGTTGATATAATCTGCGACAAGATGTCCGAAAGTCTTGATTCGGAGCCAGCGATGTTGGCCGGAGGAGCCCGTGGTCAGCGTCTCACGGAGCAGCCTCAAGGAAGCAACAATCCTAACTGTAACTGCTAA
Protein
MTGEAKWQKDAADQNFDYMFKLLIIGNSSVGKTSFLFRYADDSFTSAFVSTVGIDFKVKTVFRHDKRVKLQIWDTAGQERYRTITTAYYRGAMGFILMYDITNEESFNSVQDWVTQIKTYSWDNAQVILVGNKCDMEEERVVSAERGRQLADQLGVEFYETSAKENINVKAVFERLVDIICDKMSESLDSEPAMLAGGARGQRLTEQPQGSNNPNCNC
Summary
Description
Involved in exocytosis by regulating a late step in synaptic vesicle fusion. Could play a role in neurotransmitter release by regulating membrane flow in the nerve terminal (By similarity).
Subunit
Interacts with Rph.
Similarity
Belongs to the small GTPase superfamily. Rab family.
Keywords
3D-structure
Cell junction
Complete proteome
Cytoplasmic vesicle
Exocytosis
GTP-binding
Lipoprotein
Methylation
Nucleotide-binding
Prenylation
Protein transport
Reference proteome
Synapse
Transport
Feature
chain Ras-related protein Rab-3
Uniprot
Q19KB6
A0A3S2M535
A0A089FP72
A0A194R171
A0A1Q3FZV9
B0WTB7
+ More
W8BDQ5 Q176R5 A0A023EJZ7 A0A0L0BPK0 A0A0A1X6N8 A0A0K8W7J2 A0A034VXF6 A0A0J9R9X3 B4P843 B4HMT4 B3NN60 A0A1W4VPM8 P25228 B4MNY2 A0A0A9XQQ7 A0A1B0CFP3 A0A1I8QE87 A0A1I8N6C6 A0A1B6LNV3 A0A3B0J454 Q28XJ2 B4GG85 B3MIJ6 A0A1B6EKV0 A0A1L8DV26 A0A2R7VU72 B4KN12 A0A0Q9XEQ3 A0A1J1J029 A0A1B6MV51 A0A1B0G6P1 A0A1A9ZX66 A0A1B6HRY1 A0A1B6DB46 A0A1A9YB24 A0A1A9VY18 A0A1B0BHL4 A0A336K368 D6X1H3 A0A1B6H1V7 E0VAW9 A0A1B6C9J6 A0A069DXG1 T1HH21 A0A0M3QUG5 W5JA76 A0A3F2YSG8 E2BGB1 A0A182YBZ1 Q7PPZ7 A0A3F2YSF8 A0A2P8YMP8 A0A0T6B704 B4K0H1 B4LKN6 A0A084VUF0 A0A182J5W5 L7M483 A0A224Z9H3 A0A131Z459 L7M7Q4 A0A131XB24 A0A023GGB9 A0A2J7PDR4 J9K5D5 A0A2S2P0S0 A0A2H8TJI8 A0A182T864 A0A182V5V8 A0A182FCT5 A0A0C9R8L2 A0A182X8H7 A0A0C9QFP4 A0A182RP42 A0A182WMH0 A0A182PFZ2 A0A182LS46 A0A182PZH2 A0A182N9N0 A0A1Y1L537 V5IDU4 B7PEJ6 E2AJT5 A0A088AML5 A0A2A3E5P0 V9IG11 A0A3L8D461 A0A026W5E7 A0A0J7L3B6 A0A0L8G4F2 A0A0L7RAG5 A0A195C262 A0A2R5LNM8 A0A293M1Q6
W8BDQ5 Q176R5 A0A023EJZ7 A0A0L0BPK0 A0A0A1X6N8 A0A0K8W7J2 A0A034VXF6 A0A0J9R9X3 B4P843 B4HMT4 B3NN60 A0A1W4VPM8 P25228 B4MNY2 A0A0A9XQQ7 A0A1B0CFP3 A0A1I8QE87 A0A1I8N6C6 A0A1B6LNV3 A0A3B0J454 Q28XJ2 B4GG85 B3MIJ6 A0A1B6EKV0 A0A1L8DV26 A0A2R7VU72 B4KN12 A0A0Q9XEQ3 A0A1J1J029 A0A1B6MV51 A0A1B0G6P1 A0A1A9ZX66 A0A1B6HRY1 A0A1B6DB46 A0A1A9YB24 A0A1A9VY18 A0A1B0BHL4 A0A336K368 D6X1H3 A0A1B6H1V7 E0VAW9 A0A1B6C9J6 A0A069DXG1 T1HH21 A0A0M3QUG5 W5JA76 A0A3F2YSG8 E2BGB1 A0A182YBZ1 Q7PPZ7 A0A3F2YSF8 A0A2P8YMP8 A0A0T6B704 B4K0H1 B4LKN6 A0A084VUF0 A0A182J5W5 L7M483 A0A224Z9H3 A0A131Z459 L7M7Q4 A0A131XB24 A0A023GGB9 A0A2J7PDR4 J9K5D5 A0A2S2P0S0 A0A2H8TJI8 A0A182T864 A0A182V5V8 A0A182FCT5 A0A0C9R8L2 A0A182X8H7 A0A0C9QFP4 A0A182RP42 A0A182WMH0 A0A182PFZ2 A0A182LS46 A0A182PZH2 A0A182N9N0 A0A1Y1L537 V5IDU4 B7PEJ6 E2AJT5 A0A088AML5 A0A2A3E5P0 V9IG11 A0A3L8D461 A0A026W5E7 A0A0J7L3B6 A0A0L8G4F2 A0A0L7RAG5 A0A195C262 A0A2R5LNM8 A0A293M1Q6
Pubmed
19121390
28756777
26354079
24495485
17510324
24945155
+ More
26483478 26108605 25830018 25348373 22936249 17994087 17550304 1648935 14722103 10731132 12537572 12537569 25401762 25315136 15632085 18362917 19820115 20566863 26334808 20920257 23761445 20798317 25244985 12364791 14747013 17210077 29403074 24438588 25576852 28797301 26830274 28049606 28004739 25765539 30249741 24508170
26483478 26108605 25830018 25348373 22936249 17994087 17550304 1648935 14722103 10731132 12537572 12537569 25401762 25315136 15632085 18362917 19820115 20566863 26334808 20920257 23761445 20798317 25244985 12364791 14747013 17210077 29403074 24438588 25576852 28797301 26830274 28049606 28004739 25765539 30249741 24508170
EMBL
BABH01013209
DQ525204
ABF71469.1
RSAL01000037
RVE51147.1
KM035414
+ More
KZ149892 AIP87049.1 PZC79043.1 KQ460883 KPJ11452.1 GFDL01001972 JAV33073.1 DS232084 EDS34351.1 GAMC01011442 GAMC01011441 JAB95113.1 CH477383 EAT42176.1 JXUM01095945 JXUM01095946 JXUM01095947 GAPW01004183 KQ564233 JAC09415.1 KXJ72553.1 JRES01001567 KNC21982.1 GBXI01007303 JAD06989.1 GDHF01005156 GDHF01005114 JAI47158.1 JAI47200.1 GAKP01010942 GAKP01010941 JAC48011.1 CM002911 KMY92841.1 CM000158 EDW90090.1 CH480816 EDW47296.1 CH954179 EDV56580.1 M64621 AB112932 AE013599 AY060449 CH963848 EDW73821.1 GBHO01021638 GBRD01009196 JAG21966.1 JAG56625.1 AJWK01010224 AJWK01010225 AJWK01010226 AJWK01010227 GEBQ01020569 GEBQ01014629 GEBQ01013762 GEBQ01011838 GEBQ01008793 JAT19408.1 JAT25348.1 JAT26215.1 JAT28139.1 JAT31184.1 OUUW01000001 SPP74142.1 CM000071 EAL26324.1 CH479183 EDW35505.1 CH902619 EDV38072.1 GECZ01031205 JAS38564.1 GFDF01003790 JAV10294.1 KK854072 PTY10698.1 CH933808 EDW08839.1 KRG04298.1 CVRI01000065 CRL05813.1 GEBQ01028079 GEBQ01021572 GEBQ01013296 GEBQ01005106 GEBQ01000180 JAT11898.1 JAT18405.1 JAT26681.1 JAT34871.1 JAT39797.1 CCAG010015977 CCAG010015978 GECU01030296 JAS77410.1 GEDC01014473 JAS22825.1 JXJN01014427 JXJN01014428 UFQS01000063 UFQT01000063 SSW98861.1 SSX19247.1 KQ971371 EFA09496.2 GECZ01001093 JAS68676.1 DS235019 EEB10525.1 GEDC01027131 JAS10167.1 GBGD01002845 JAC86044.1 ACPB03001028 CP012524 ALC40623.1 ADMH02002051 ETN59755.1 APCN01002761 GL448138 EFN85274.1 AAAB01008900 EAA09493.2 PYGN01000487 PSN45521.1 LJIG01009425 KRT83106.1 CH916446 EDV90871.1 CH940648 EDW61759.1 ATLV01016737 KE525103 KFB41594.1 AXCP01004228 GACK01005918 JAA59116.1 GFPF01012198 MAA23344.1 GEDV01002393 JAP86164.1 GACK01005905 JAA59129.1 GEFH01004834 JAP63747.1 GBBM01002446 JAC32972.1 NEVH01026385 PNF14468.1 ABLF02017393 GGMR01010189 MBY22808.1 GFXV01001643 MBW13448.1 GBYB01003131 JAG72898.1 GBYB01013303 JAG83070.1 AXCM01012845 AXCM01012846 AXCN02001010 GEZM01066380 GEZM01066379 JAV67918.1 GANP01011332 JAB73136.1 ABJB010261944 ABJB010797507 DS695319 EEC05018.1 GL440100 EFN66251.1 KZ288361 PBC27015.1 JR040728 AEY59264.1 QOIP01000013 RLU15292.1 KK107467 EZA50254.1 LBMM01001103 KMQ96919.1 KQ423910 KOF71922.1 KQ414618 KOC67892.1 KQ978344 KYM94959.1 GGLE01006985 MBY11111.1 GFWV01009239 MAA33968.1
KZ149892 AIP87049.1 PZC79043.1 KQ460883 KPJ11452.1 GFDL01001972 JAV33073.1 DS232084 EDS34351.1 GAMC01011442 GAMC01011441 JAB95113.1 CH477383 EAT42176.1 JXUM01095945 JXUM01095946 JXUM01095947 GAPW01004183 KQ564233 JAC09415.1 KXJ72553.1 JRES01001567 KNC21982.1 GBXI01007303 JAD06989.1 GDHF01005156 GDHF01005114 JAI47158.1 JAI47200.1 GAKP01010942 GAKP01010941 JAC48011.1 CM002911 KMY92841.1 CM000158 EDW90090.1 CH480816 EDW47296.1 CH954179 EDV56580.1 M64621 AB112932 AE013599 AY060449 CH963848 EDW73821.1 GBHO01021638 GBRD01009196 JAG21966.1 JAG56625.1 AJWK01010224 AJWK01010225 AJWK01010226 AJWK01010227 GEBQ01020569 GEBQ01014629 GEBQ01013762 GEBQ01011838 GEBQ01008793 JAT19408.1 JAT25348.1 JAT26215.1 JAT28139.1 JAT31184.1 OUUW01000001 SPP74142.1 CM000071 EAL26324.1 CH479183 EDW35505.1 CH902619 EDV38072.1 GECZ01031205 JAS38564.1 GFDF01003790 JAV10294.1 KK854072 PTY10698.1 CH933808 EDW08839.1 KRG04298.1 CVRI01000065 CRL05813.1 GEBQ01028079 GEBQ01021572 GEBQ01013296 GEBQ01005106 GEBQ01000180 JAT11898.1 JAT18405.1 JAT26681.1 JAT34871.1 JAT39797.1 CCAG010015977 CCAG010015978 GECU01030296 JAS77410.1 GEDC01014473 JAS22825.1 JXJN01014427 JXJN01014428 UFQS01000063 UFQT01000063 SSW98861.1 SSX19247.1 KQ971371 EFA09496.2 GECZ01001093 JAS68676.1 DS235019 EEB10525.1 GEDC01027131 JAS10167.1 GBGD01002845 JAC86044.1 ACPB03001028 CP012524 ALC40623.1 ADMH02002051 ETN59755.1 APCN01002761 GL448138 EFN85274.1 AAAB01008900 EAA09493.2 PYGN01000487 PSN45521.1 LJIG01009425 KRT83106.1 CH916446 EDV90871.1 CH940648 EDW61759.1 ATLV01016737 KE525103 KFB41594.1 AXCP01004228 GACK01005918 JAA59116.1 GFPF01012198 MAA23344.1 GEDV01002393 JAP86164.1 GACK01005905 JAA59129.1 GEFH01004834 JAP63747.1 GBBM01002446 JAC32972.1 NEVH01026385 PNF14468.1 ABLF02017393 GGMR01010189 MBY22808.1 GFXV01001643 MBW13448.1 GBYB01003131 JAG72898.1 GBYB01013303 JAG83070.1 AXCM01012845 AXCM01012846 AXCN02001010 GEZM01066380 GEZM01066379 JAV67918.1 GANP01011332 JAB73136.1 ABJB010261944 ABJB010797507 DS695319 EEC05018.1 GL440100 EFN66251.1 KZ288361 PBC27015.1 JR040728 AEY59264.1 QOIP01000013 RLU15292.1 KK107467 EZA50254.1 LBMM01001103 KMQ96919.1 KQ423910 KOF71922.1 KQ414618 KOC67892.1 KQ978344 KYM94959.1 GGLE01006985 MBY11111.1 GFWV01009239 MAA33968.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000002320
UP000008820
UP000069940
+ More
UP000249989 UP000037069 UP000002282 UP000001292 UP000008711 UP000192221 UP000000803 UP000007798 UP000092461 UP000095300 UP000095301 UP000268350 UP000001819 UP000008744 UP000007801 UP000009192 UP000183832 UP000092444 UP000092445 UP000092443 UP000078200 UP000092460 UP000007266 UP000009046 UP000015103 UP000092553 UP000000673 UP000075840 UP000008237 UP000076408 UP000007062 UP000245037 UP000001070 UP000008792 UP000030765 UP000075880 UP000235965 UP000007819 UP000075901 UP000075903 UP000069272 UP000076407 UP000075900 UP000075920 UP000075885 UP000075883 UP000075886 UP000075884 UP000001555 UP000000311 UP000005203 UP000242457 UP000279307 UP000053097 UP000036403 UP000053454 UP000053825 UP000078542
UP000249989 UP000037069 UP000002282 UP000001292 UP000008711 UP000192221 UP000000803 UP000007798 UP000092461 UP000095300 UP000095301 UP000268350 UP000001819 UP000008744 UP000007801 UP000009192 UP000183832 UP000092444 UP000092445 UP000092443 UP000078200 UP000092460 UP000007266 UP000009046 UP000015103 UP000092553 UP000000673 UP000075840 UP000008237 UP000076408 UP000007062 UP000245037 UP000001070 UP000008792 UP000030765 UP000075880 UP000235965 UP000007819 UP000075901 UP000075903 UP000069272 UP000076407 UP000075900 UP000075920 UP000075885 UP000075883 UP000075886 UP000075884 UP000001555 UP000000311 UP000005203 UP000242457 UP000279307 UP000053097 UP000036403 UP000053454 UP000053825 UP000078542
Interpro
CDD
ProteinModelPortal
Q19KB6
A0A3S2M535
A0A089FP72
A0A194R171
A0A1Q3FZV9
B0WTB7
+ More
W8BDQ5 Q176R5 A0A023EJZ7 A0A0L0BPK0 A0A0A1X6N8 A0A0K8W7J2 A0A034VXF6 A0A0J9R9X3 B4P843 B4HMT4 B3NN60 A0A1W4VPM8 P25228 B4MNY2 A0A0A9XQQ7 A0A1B0CFP3 A0A1I8QE87 A0A1I8N6C6 A0A1B6LNV3 A0A3B0J454 Q28XJ2 B4GG85 B3MIJ6 A0A1B6EKV0 A0A1L8DV26 A0A2R7VU72 B4KN12 A0A0Q9XEQ3 A0A1J1J029 A0A1B6MV51 A0A1B0G6P1 A0A1A9ZX66 A0A1B6HRY1 A0A1B6DB46 A0A1A9YB24 A0A1A9VY18 A0A1B0BHL4 A0A336K368 D6X1H3 A0A1B6H1V7 E0VAW9 A0A1B6C9J6 A0A069DXG1 T1HH21 A0A0M3QUG5 W5JA76 A0A3F2YSG8 E2BGB1 A0A182YBZ1 Q7PPZ7 A0A3F2YSF8 A0A2P8YMP8 A0A0T6B704 B4K0H1 B4LKN6 A0A084VUF0 A0A182J5W5 L7M483 A0A224Z9H3 A0A131Z459 L7M7Q4 A0A131XB24 A0A023GGB9 A0A2J7PDR4 J9K5D5 A0A2S2P0S0 A0A2H8TJI8 A0A182T864 A0A182V5V8 A0A182FCT5 A0A0C9R8L2 A0A182X8H7 A0A0C9QFP4 A0A182RP42 A0A182WMH0 A0A182PFZ2 A0A182LS46 A0A182PZH2 A0A182N9N0 A0A1Y1L537 V5IDU4 B7PEJ6 E2AJT5 A0A088AML5 A0A2A3E5P0 V9IG11 A0A3L8D461 A0A026W5E7 A0A0J7L3B6 A0A0L8G4F2 A0A0L7RAG5 A0A195C262 A0A2R5LNM8 A0A293M1Q6
W8BDQ5 Q176R5 A0A023EJZ7 A0A0L0BPK0 A0A0A1X6N8 A0A0K8W7J2 A0A034VXF6 A0A0J9R9X3 B4P843 B4HMT4 B3NN60 A0A1W4VPM8 P25228 B4MNY2 A0A0A9XQQ7 A0A1B0CFP3 A0A1I8QE87 A0A1I8N6C6 A0A1B6LNV3 A0A3B0J454 Q28XJ2 B4GG85 B3MIJ6 A0A1B6EKV0 A0A1L8DV26 A0A2R7VU72 B4KN12 A0A0Q9XEQ3 A0A1J1J029 A0A1B6MV51 A0A1B0G6P1 A0A1A9ZX66 A0A1B6HRY1 A0A1B6DB46 A0A1A9YB24 A0A1A9VY18 A0A1B0BHL4 A0A336K368 D6X1H3 A0A1B6H1V7 E0VAW9 A0A1B6C9J6 A0A069DXG1 T1HH21 A0A0M3QUG5 W5JA76 A0A3F2YSG8 E2BGB1 A0A182YBZ1 Q7PPZ7 A0A3F2YSF8 A0A2P8YMP8 A0A0T6B704 B4K0H1 B4LKN6 A0A084VUF0 A0A182J5W5 L7M483 A0A224Z9H3 A0A131Z459 L7M7Q4 A0A131XB24 A0A023GGB9 A0A2J7PDR4 J9K5D5 A0A2S2P0S0 A0A2H8TJI8 A0A182T864 A0A182V5V8 A0A182FCT5 A0A0C9R8L2 A0A182X8H7 A0A0C9QFP4 A0A182RP42 A0A182WMH0 A0A182PFZ2 A0A182LS46 A0A182PZH2 A0A182N9N0 A0A1Y1L537 V5IDU4 B7PEJ6 E2AJT5 A0A088AML5 A0A2A3E5P0 V9IG11 A0A3L8D461 A0A026W5E7 A0A0J7L3B6 A0A0L8G4F2 A0A0L7RAG5 A0A195C262 A0A2R5LNM8 A0A293M1Q6
PDB
4RKF
E-value=3.67673e-96,
Score=894
Ontologies
GO
GO:0003924
GO:0005525
GO:0048789
GO:0008021
GO:0048790
GO:0048786
GO:0016192
GO:0030054
GO:0045202
GO:0031982
GO:0032482
GO:0007269
GO:0031630
GO:0006887
GO:0048172
GO:0015031
GO:0005768
GO:0009306
GO:0006886
GO:0017157
GO:0006904
GO:0072659
GO:0005886
GO:0008168
GO:0004767
GO:0007264
GO:0006468
GO:0004683
GO:0005516
GO:0005515
GO:0008237
GO:0008152
GO:0016491
GO:0015930
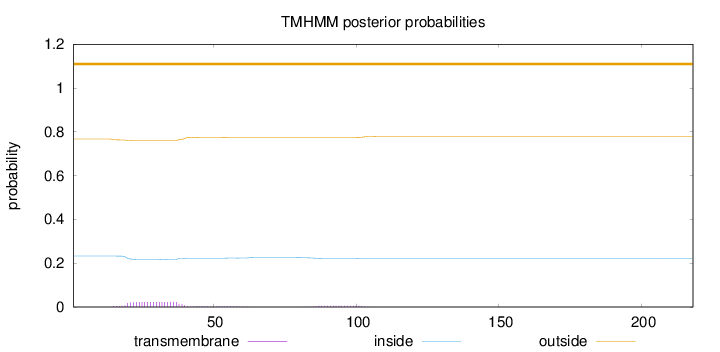
Topology
Subcellular location
Length:
218
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.61879
Exp number, first 60 AAs:
0.51826
Total prob of N-in:
0.23297
outside
1 - 218
Population Genetic Test Statistics
Pi
300.365318
Theta
213.012135
Tajima's D
1.049488
CLR
0.851609
CSRT
0.671816409179541
Interpretation
Uncertain
