Pre Gene Modal
BGIBMGA000408
Annotation
Ca2_/calmodulin-dependent_protein_kinase_II_[Bombyx_mori]
Full name
Calcium/calmodulin-dependent protein kinase type II alpha chain
Location in the cell
Cytoplasmic Reliability : 2.103 Nuclear Reliability : 1.978
Sequence
CDS
ATGGCTAATCCAAATCGTGAAAGTGTTAGCACGCGGTTTTCGGACAACTATGAACTTAAGGAAGAATTAGGCAAGGGCGCCTTCTCGATTGTTAGGAGAGCTGTACAAAAGTCTACTGGATACGAGTTTGCGGCAAAAATCATAAATACCAAGAAACTCTCTGCAAGAGACTTCCAGAAATTGGAACGAGAAGCAAGAATCTGTCGTAAACTACAGCATCCCAATATTGTGCGACTACATGATTCCATTCAAGAAGAACATTGCCATTACCTTGTCTTTGATTTGGTTACCGGGGGTGAATTATTTGAAGACATAGTGGCGCGAGAATTTTATTCTGAAGCTGATGCGTCACACTGCATTCAGCAAATATTAGAATCTGTCCATCACTGTCACCATAATGGAGTTGTGCACAGAGATCTGAAACCAGAAAATCTTTTGCTCGCAAGCAAAGCTAAGGGAGCTGCTGTCAAGTTGGCCGATTTTGGGTTAGCTATCGAAGTTCAAGGAGATCAACAAGCATGGTTCGGATTCGCAGGTACACCCGGTTACTTATCGCCTGAAGTATTAAAAAAGGAACCTTATGGTAAACCAGTGGATATATGGGCTTGTGGGGTCATTCTATACATATTGTTAGTTGGCTACCCTCCATTTTGGGATGAAGATCAGTATCGTCTCTATGCGCAAATCAAAGCGGGAGCATATGATTATCCGTCTCCTGAGTGGGATACCGTTACTCCTGAAGCTAAGAGCCTTATTAATCAAATGCTTACTGTTAATCCTAGCAAAAGGATCACCGCTTCCGAAGCTCTGAAACATCCATGGATTTGCCACCGGGAACGCGTTGCTTCAGTTATGCATAGACAGGAAACTGTGGATTGTTTGAAAAAATTCAATGCACGTCGCAAGCTCAAGGGTGCAATTTTAACAACGATGCTTGCTACCAGAAATTTCTCTGGTAAATCAATGGTTAATAAAAAAGGCGATGGATCGCAAGTAAAAGAGTCTACTGATAGTAGTACAACGCTTGAAGATGATGATTTAGACAAAGACAAGAAAGGAGTGGACCGAGCATGCACGGTTATATCTAAAGAACATGACGAAGAAGCCCTGACAAAAGCGGACTCGTTTGGAAAAGCTCGGGGCGATAGTAATGCTTCTTTGCGCCGAGCGGAAGTGATTAAGGTTACTGAGGTGCTGATAGATGCAATCAATAATGGTGATTACGACACATATTCCAAGCTATGTGATCCCAATGTGACAGCTTTTGATCCTGATGCTCTAGGAAATCTGATTGAAGGTGTAGAATTCCATAAATTCTTTATTGACAATACACCGCCACATGTGAAAACTAATACCACTATTCTGAATCCAAGAGTACATCTTCTCGGTGATGATGTTGCAGTCATAGCATACGTCTGCGTAACACAAAGCGTAGATTCCGAAGGTCGACGGGCTACACACCAATCACAAGAGACTCGCATTTGGCACAAACGTCACAACAAGTGGACTGCCGTTCATTTCCATCGCTCCTAA
Protein
MANPNRESVSTRFSDNYELKEELGKGAFSIVRRAVQKSTGYEFAAKIINTKKLSARDFQKLEREARICRKLQHPNIVRLHDSIQEEHCHYLVFDLVTGGELFEDIVAREFYSEADASHCIQQILESVHHCHHNGVVHRDLKPENLLLASKAKGAAVKLADFGLAIEVQGDQQAWFGFAGTPGYLSPEVLKKEPYGKPVDIWACGVILYILLVGYPPFWDEDQYRLYAQIKAGAYDYPSPEWDTVTPEAKSLINQMLTVNPSKRITASEALKHPWICHRERVASVMHRQETVDCLKKFNARRKLKGAILTTMLATRNFSGKSMVNKKGDGSQVKESTDSSTTLEDDDLDKDKKGVDRACTVISKEHDEEALTKADSFGKARGDSNASLRRAEVIKVTEVLIDAINNGDYDTYSKLCDPNVTAFDPDALGNLIEGVEFHKFFIDNTPPHVKTNTTILNPRVHLLGDDVAVIAYVCVTQSVDSEGRRATHQSQETRIWHKRHNKWTAVHFHRS
Summary
Description
A key regulator of plasticity in synaptic physiology and behavior, alterations in its activity produce pleiotrophic effects that involve synaptic transmission and development as well as various aspects of behavior. Directly modulates eag potassium channels.
Catalytic Activity
ATP + L-seryl-[protein] = ADP + H(+) + O-phospho-L-seryl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
ATP + L-threonyl-[protein] = ADP + H(+) + O-phospho-L-threonyl-[protein]
Subunit
Interacts with CASK.
Similarity
Belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family. CaMK subfamily.
Keywords
3D-structure
Alternative splicing
ATP-binding
Calmodulin-binding
Complete proteome
Kinase
Nucleotide-binding
Phosphoprotein
Reference proteome
Transferase
Feature
chain Calcium/calmodulin-dependent protein kinase type II alpha chain
splice variant In isoform 4.
splice variant In isoform 4.
Uniprot
D6RVR8
A0A2A4IUJ3
A0A2H1VUL8
A0A2W1C002
A0A068ETN5
A0A194R1R1
+ More
A0A194PXS1 A0A212EQX4 A0A0L7KU84 L0MLR4 M9T9J6 A0A1W4VIE5 A0A1W4V592 A0A0Q5SUI1 H6SWV1 A0A0Q5SQ34 A0A0J9S2S8 A0A1W4VHL9 A0A0J9S152 A0A3B0K408 B4H9P5 D6WFJ6 B3MY38 A0A0P8XJA8 A0A0Q5SGF2 A0A3B0K6M9 K7IZZ8 E9GTA8 A0A1W4V4T2 A0A1J1J8D8 D1YSG7 B3P9P9 Q00168 A0A0P8XZJ0 A0A0J9S143 W8BK87 D1YSG8 B4NHL4 B4IKQ9 A0A0J9S2U8 A0A0A1X1U5 A0A0P9A9G3 B4L7C9 A0A158NYG2 A0A0Q9WUI9 B4JZN8 A0A0P6HFA1 B4MEZ2 A0A0M5J0P2 A0A0Q9XG85 A0A1B6M762 D0Z795 A0A1B6IH83 Q00168-2 A0A1W4VIF0 A0A034VW58 A0A1L8DKE5 A0A1W4V4U2 H6SWV0 A0A0Q5SUX1 A0A0J9UST1 A0A0P8ZJH8 A0A1W4VHM5 A0A0J9S1B9 A0A3B0K460 A0A0P9BTH1 A0A0P5IZC6 A0A0Q5SKE6 A0A1W4V597 A0A3B0K8F2 L0MLP2 A0A1L8DKH0 A0A0Q5SDU8 Q00168-3 A0A0J9S149 A4V134 A0A0A1WE61 A0A2I6SFN2 A0A0Q9XNP1 A0A034VR42 A0A1W4V4T9 A0A1W4VH12 A0A0Q9WKK4 A0A0J9USU2 A0A232FAJ7 A0A0P9BTI4 A0A0Q5SRX4 W8C8L1 A0A0J9UST8 A0A0Q5SDZ8 A0A0P6GF02 A0A0J9S1C8 T1INT6 A0A1J1J5E5 A0A0Q9XI62 A0A0Q9XFD3 A0A0P5SXQ1 A0A0P4WI42 A0A0A9X1X9 A0A131XKK0
A0A194PXS1 A0A212EQX4 A0A0L7KU84 L0MLR4 M9T9J6 A0A1W4VIE5 A0A1W4V592 A0A0Q5SUI1 H6SWV1 A0A0Q5SQ34 A0A0J9S2S8 A0A1W4VHL9 A0A0J9S152 A0A3B0K408 B4H9P5 D6WFJ6 B3MY38 A0A0P8XJA8 A0A0Q5SGF2 A0A3B0K6M9 K7IZZ8 E9GTA8 A0A1W4V4T2 A0A1J1J8D8 D1YSG7 B3P9P9 Q00168 A0A0P8XZJ0 A0A0J9S143 W8BK87 D1YSG8 B4NHL4 B4IKQ9 A0A0J9S2U8 A0A0A1X1U5 A0A0P9A9G3 B4L7C9 A0A158NYG2 A0A0Q9WUI9 B4JZN8 A0A0P6HFA1 B4MEZ2 A0A0M5J0P2 A0A0Q9XG85 A0A1B6M762 D0Z795 A0A1B6IH83 Q00168-2 A0A1W4VIF0 A0A034VW58 A0A1L8DKE5 A0A1W4V4U2 H6SWV0 A0A0Q5SUX1 A0A0J9UST1 A0A0P8ZJH8 A0A1W4VHM5 A0A0J9S1B9 A0A3B0K460 A0A0P9BTH1 A0A0P5IZC6 A0A0Q5SKE6 A0A1W4V597 A0A3B0K8F2 L0MLP2 A0A1L8DKH0 A0A0Q5SDU8 Q00168-3 A0A0J9S149 A4V134 A0A0A1WE61 A0A2I6SFN2 A0A0Q9XNP1 A0A034VR42 A0A1W4V4T9 A0A1W4VH12 A0A0Q9WKK4 A0A0J9USU2 A0A232FAJ7 A0A0P9BTI4 A0A0Q5SRX4 W8C8L1 A0A0J9UST8 A0A0Q5SDZ8 A0A0P6GF02 A0A0J9S1C8 T1INT6 A0A1J1J5E5 A0A0Q9XI62 A0A0Q9XFD3 A0A0P5SXQ1 A0A0P4WI42 A0A0A9X1X9 A0A131XKK0
EC Number
2.7.11.17
Pubmed
19121390
28756777
26354079
22118469
26227816
10731132
+ More
12537568 12537572 12537573 12537574 16110336 17569856 17569867 23740573 17994087 22936249 18362917 19820115 20075255 21292972 8380587 1910789 8397298 11980904 14687552 16880127 18327897 24495485 25830018 18057021 21347285 20479145 25348373 28648823 25401762 26823975 28049606
12537568 12537572 12537573 12537574 16110336 17569856 17569867 23740573 17994087 22936249 18362917 19820115 20075255 21292972 8380587 1910789 8397298 11980904 14687552 16880127 18327897 24495485 25830018 18057021 21347285 20479145 25348373 28648823 25401762 26823975 28049606
EMBL
BABH01013256
AB563704
BAJ08364.1
NWSH01007804
PCG62803.1
ODYU01004251
+ More
SOQ43924.1 KZ149892 PZC79035.1 KJ650044 AID54518.1 KQ460883 KPJ11459.1 KQ459586 KPI97823.1 AGBW02013205 OWR43869.1 JTDY01005762 KOB66660.1 AE014135 AGB96573.1 KC733178 AGI98593.1 CH954184 KQS21460.1 HQ176457 ADX05544.1 KQS21461.1 CM002916 KMZ07355.1 KMZ07359.1 OUUW01000017 SPP88944.1 CH479230 EDW36529.1 KQ971328 EEZ99807.2 CH902631 EDV34070.1 KPU74954.1 KQS21456.1 SPP88943.1 GL732563 EFX77393.1 CVRI01000070 CRL07177.1 ACZ95101.1 EDV45212.1 D13330 D13331 D13332 D13333 M74583 S65712 S65716 S65717 S65719 S65724 KPU74953.1 KMZ07357.1 GAMC01007408 JAB99147.1 ACZ95102.1 CH964272 EDW83583.1 CH480855 EDW52649.1 KMZ07353.1 KMZ07361.1 KMZ07363.1 KMZ07364.1 KMZ07370.1 GBXI01009887 GBXI01009476 GBXI01000596 JAD04405.1 JAD04816.1 JAD13696.1 KPU74955.1 KPU74956.1 KPU74957.1 CH933813 EDW10923.1 ADTU01003797 ADTU01003798 ADTU01003799 ADTU01003800 ADTU01003801 ADTU01003802 ADTU01003803 ADTU01003804 ADTU01003805 ADTU01003806 KRF99822.1 CH916380 EDV90904.1 GDIQ01029462 JAN65275.1 CH940665 EDW71093.1 CP012527 ALC48081.1 KRG07285.1 GEBQ01008219 JAT31758.1 CM000831 ACY70533.1 KRF85336.1 KRF85339.1 GECU01021431 JAS86275.1 GAKP01013220 GAKP01013219 GAKP01013210 JAC45733.1 GFDF01007141 JAV06943.1 HQ176456 ADX05543.1 KQS21462.1 KMZ07356.1 KPU74952.1 KMZ07358.1 SPP88945.1 KPU74951.1 GDIQ01205404 GDIQ01203754 GDIP01045962 JAK46321.1 JAM57753.1 KQS21457.1 SPP88942.1 AGB96574.1 GFDF01007142 JAV06942.1 KQS21459.1 KMZ07362.1 AAN06569.2 GBXI01017316 JAC96975.1 KY552652 AUO16381.1 KRG07290.1 GAKP01013218 GAKP01013211 JAC45741.1 KRF85341.1 KMZ07371.1 NNAY01000525 OXU27866.1 KPU74960.1 KQS21454.1 GAMC01007406 JAB99149.1 KMZ07366.1 KQS21458.1 GDIQ01035859 JAN58878.1 KMZ07368.1 JH431210 CRL07178.1 KRG07289.1 KRG07292.1 GDIQ01241340 GDIP01136672 JAK10385.1 JAL67042.1 GDRN01047452 JAI66722.1 GBHO01028877 GDHC01001256 JAG14727.1 JAQ17373.1 GEFH01001856 JAP66725.1
SOQ43924.1 KZ149892 PZC79035.1 KJ650044 AID54518.1 KQ460883 KPJ11459.1 KQ459586 KPI97823.1 AGBW02013205 OWR43869.1 JTDY01005762 KOB66660.1 AE014135 AGB96573.1 KC733178 AGI98593.1 CH954184 KQS21460.1 HQ176457 ADX05544.1 KQS21461.1 CM002916 KMZ07355.1 KMZ07359.1 OUUW01000017 SPP88944.1 CH479230 EDW36529.1 KQ971328 EEZ99807.2 CH902631 EDV34070.1 KPU74954.1 KQS21456.1 SPP88943.1 GL732563 EFX77393.1 CVRI01000070 CRL07177.1 ACZ95101.1 EDV45212.1 D13330 D13331 D13332 D13333 M74583 S65712 S65716 S65717 S65719 S65724 KPU74953.1 KMZ07357.1 GAMC01007408 JAB99147.1 ACZ95102.1 CH964272 EDW83583.1 CH480855 EDW52649.1 KMZ07353.1 KMZ07361.1 KMZ07363.1 KMZ07364.1 KMZ07370.1 GBXI01009887 GBXI01009476 GBXI01000596 JAD04405.1 JAD04816.1 JAD13696.1 KPU74955.1 KPU74956.1 KPU74957.1 CH933813 EDW10923.1 ADTU01003797 ADTU01003798 ADTU01003799 ADTU01003800 ADTU01003801 ADTU01003802 ADTU01003803 ADTU01003804 ADTU01003805 ADTU01003806 KRF99822.1 CH916380 EDV90904.1 GDIQ01029462 JAN65275.1 CH940665 EDW71093.1 CP012527 ALC48081.1 KRG07285.1 GEBQ01008219 JAT31758.1 CM000831 ACY70533.1 KRF85336.1 KRF85339.1 GECU01021431 JAS86275.1 GAKP01013220 GAKP01013219 GAKP01013210 JAC45733.1 GFDF01007141 JAV06943.1 HQ176456 ADX05543.1 KQS21462.1 KMZ07356.1 KPU74952.1 KMZ07358.1 SPP88945.1 KPU74951.1 GDIQ01205404 GDIQ01203754 GDIP01045962 JAK46321.1 JAM57753.1 KQS21457.1 SPP88942.1 AGB96574.1 GFDF01007142 JAV06942.1 KQS21459.1 KMZ07362.1 AAN06569.2 GBXI01017316 JAC96975.1 KY552652 AUO16381.1 KRG07290.1 GAKP01013218 GAKP01013211 JAC45741.1 KRF85341.1 KMZ07371.1 NNAY01000525 OXU27866.1 KPU74960.1 KQS21454.1 GAMC01007406 JAB99149.1 KMZ07366.1 KQS21458.1 GDIQ01035859 JAN58878.1 KMZ07368.1 JH431210 CRL07178.1 KRG07289.1 KRG07292.1 GDIQ01241340 GDIP01136672 JAK10385.1 JAL67042.1 GDRN01047452 JAI66722.1 GBHO01028877 GDHC01001256 JAG14727.1 JAQ17373.1 GEFH01001856 JAP66725.1
Proteomes
Interpro
ProteinModelPortal
D6RVR8
A0A2A4IUJ3
A0A2H1VUL8
A0A2W1C002
A0A068ETN5
A0A194R1R1
+ More
A0A194PXS1 A0A212EQX4 A0A0L7KU84 L0MLR4 M9T9J6 A0A1W4VIE5 A0A1W4V592 A0A0Q5SUI1 H6SWV1 A0A0Q5SQ34 A0A0J9S2S8 A0A1W4VHL9 A0A0J9S152 A0A3B0K408 B4H9P5 D6WFJ6 B3MY38 A0A0P8XJA8 A0A0Q5SGF2 A0A3B0K6M9 K7IZZ8 E9GTA8 A0A1W4V4T2 A0A1J1J8D8 D1YSG7 B3P9P9 Q00168 A0A0P8XZJ0 A0A0J9S143 W8BK87 D1YSG8 B4NHL4 B4IKQ9 A0A0J9S2U8 A0A0A1X1U5 A0A0P9A9G3 B4L7C9 A0A158NYG2 A0A0Q9WUI9 B4JZN8 A0A0P6HFA1 B4MEZ2 A0A0M5J0P2 A0A0Q9XG85 A0A1B6M762 D0Z795 A0A1B6IH83 Q00168-2 A0A1W4VIF0 A0A034VW58 A0A1L8DKE5 A0A1W4V4U2 H6SWV0 A0A0Q5SUX1 A0A0J9UST1 A0A0P8ZJH8 A0A1W4VHM5 A0A0J9S1B9 A0A3B0K460 A0A0P9BTH1 A0A0P5IZC6 A0A0Q5SKE6 A0A1W4V597 A0A3B0K8F2 L0MLP2 A0A1L8DKH0 A0A0Q5SDU8 Q00168-3 A0A0J9S149 A4V134 A0A0A1WE61 A0A2I6SFN2 A0A0Q9XNP1 A0A034VR42 A0A1W4V4T9 A0A1W4VH12 A0A0Q9WKK4 A0A0J9USU2 A0A232FAJ7 A0A0P9BTI4 A0A0Q5SRX4 W8C8L1 A0A0J9UST8 A0A0Q5SDZ8 A0A0P6GF02 A0A0J9S1C8 T1INT6 A0A1J1J5E5 A0A0Q9XI62 A0A0Q9XFD3 A0A0P5SXQ1 A0A0P4WI42 A0A0A9X1X9 A0A131XKK0
A0A194PXS1 A0A212EQX4 A0A0L7KU84 L0MLR4 M9T9J6 A0A1W4VIE5 A0A1W4V592 A0A0Q5SUI1 H6SWV1 A0A0Q5SQ34 A0A0J9S2S8 A0A1W4VHL9 A0A0J9S152 A0A3B0K408 B4H9P5 D6WFJ6 B3MY38 A0A0P8XJA8 A0A0Q5SGF2 A0A3B0K6M9 K7IZZ8 E9GTA8 A0A1W4V4T2 A0A1J1J8D8 D1YSG7 B3P9P9 Q00168 A0A0P8XZJ0 A0A0J9S143 W8BK87 D1YSG8 B4NHL4 B4IKQ9 A0A0J9S2U8 A0A0A1X1U5 A0A0P9A9G3 B4L7C9 A0A158NYG2 A0A0Q9WUI9 B4JZN8 A0A0P6HFA1 B4MEZ2 A0A0M5J0P2 A0A0Q9XG85 A0A1B6M762 D0Z795 A0A1B6IH83 Q00168-2 A0A1W4VIF0 A0A034VW58 A0A1L8DKE5 A0A1W4V4U2 H6SWV0 A0A0Q5SUX1 A0A0J9UST1 A0A0P8ZJH8 A0A1W4VHM5 A0A0J9S1B9 A0A3B0K460 A0A0P9BTH1 A0A0P5IZC6 A0A0Q5SKE6 A0A1W4V597 A0A3B0K8F2 L0MLP2 A0A1L8DKH0 A0A0Q5SDU8 Q00168-3 A0A0J9S149 A4V134 A0A0A1WE61 A0A2I6SFN2 A0A0Q9XNP1 A0A034VR42 A0A1W4V4T9 A0A1W4VH12 A0A0Q9WKK4 A0A0J9USU2 A0A232FAJ7 A0A0P9BTI4 A0A0Q5SRX4 W8C8L1 A0A0J9UST8 A0A0Q5SDZ8 A0A0P6GF02 A0A0J9S1C8 T1INT6 A0A1J1J5E5 A0A0Q9XI62 A0A0Q9XFD3 A0A0P5SXQ1 A0A0P4WI42 A0A0A9X1X9 A0A131XKK0
PDB
5U6Y
E-value=0,
Score=1826
Ontologies
PATHWAY
GO
GO:0004683
GO:0005524
GO:0005516
GO:0043005
GO:0005737
GO:0045211
GO:0048786
GO:0050710
GO:0005954
GO:0010888
GO:0030424
GO:0007611
GO:0030425
GO:0060278
GO:0004674
GO:0008049
GO:0007616
GO:0008582
GO:0051489
GO:0009267
GO:0007268
GO:0006468
GO:0007619
GO:0007528
GO:1990443
GO:0004672
GO:0005515
GO:0008237
GO:0008152
GO:0016491
GO:0015930
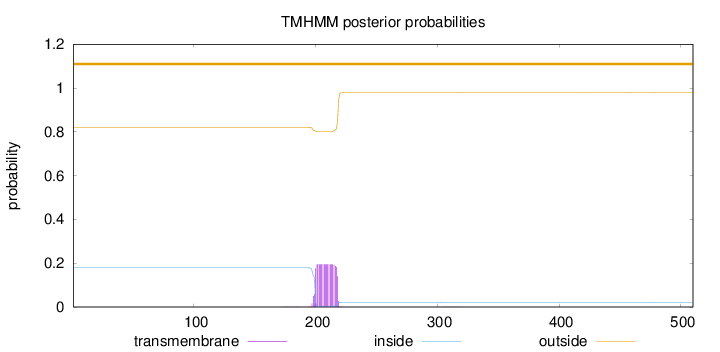
Topology
Length:
510
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.77785
Exp number, first 60 AAs:
0
Total prob of N-in:
0.17946
outside
1 - 510
Population Genetic Test Statistics
Pi
245.904893
Theta
218.404218
Tajima's D
0.420046
CLR
0.667881
CSRT
0.497275136243188
Interpretation
Uncertain
