Gene
KWMTBOMO13051 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000411
Annotation
PREDICTED:_pancreatic_lipase-related_protein_2-like_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.251
Sequence
CDS
ATGATGTACAATCGAACGAGTGCAACGTTGATGCTGCAGACCATCATGCTCTTGATGAAGAAGCAGAATTCGACGGATGTCACTAATTTCCCTGCAGGATTTAGTGATGATATGGAACTGGAGCGATGTTATGACATCTACGGATGTTTTTCTAAAGCCTACCCGTGGACGGAGCACAGACCCGATAATTATTTCCCTGCATCTGTCGAGTCAATGGCAATACGGTATCCAACTTTTACTCGTCGGAACCGAAAAATACCAGTGTTGCTTCATGCCGATAATACGGACAGAATACAAAATGCCAATCTTGATGCGAATGGACCGTTTTACTTCATATCGCACGGCTTCTTGGAAGGAGGTCACAAGAAATGGATTGAAAATCTAGCAAATGCCCTCTTAGACATTGACGGTAACGAATCAGCCACAGTGATCATAGTTGACTGGCGTGGGGGTTCCCAACCTCCGTATGGACAATCGGTTGCTAATATTAGATTGGTCGGAGTTATGACCGCCCACTTGATACACAATATTTATGAAGTTTTGCAGTTGCCACATATAGATAATTTTCATTTCATTGGTCATTCACTCGGCGCCCATTTAGGCGGTTATTGTGGCCACGAATTGCAGAAGAAGTTCAATTTGAAACTAGGACGAATAACTGGTTTGGATCCGGCCGCCCCGTACTTCAGCAGGACGGTGACTTTGGTTCGCTTGGACAGATCTGATGCTAAATATGTCGATATCATTCACAGCAACGCGATGCCTCTTTATTTCTCAGGTTTCGGAATATCAGAACCGATAGGCCACGTGGATTTCTATCCAAATGGCGGTTCGACCCAGCCCGGTTGCAAGCAATCTGACTCGCCCAACCAAAGCGCTTCGTACCAACAAGTCGTCAAGTACGTAGGCTGCAACCACGAAAGAAGCCACGAGTACTTCACGGAAAGTATTGCACCCACGTGTCCCTTCATGGCGATACAATGTGAATCTCATGAAGCGTTCCTAGCTGGCAATTGCACAAACTGCGACAACAAACACTTCTGTATACCAATGGGATATCACTCGTATGACTTATACAAACATCTTCAAATAGGCGGGCTCGTCGATACGAATACTAATATAGCTCTTTATTCGCTAACCGGTAACGCGAAACCTTATTGCAAAGTACATTACAAAGTCGTTTTGAAAGTAGCGAATTCGACGGAGAGTAAGAATCACGGACCGGAATCCGGTAAGGTAACGATCACTTTGGTGGACAGAAACAATAATAAAACCGATCGGAGATCCCTCAATCCGGAACAAAAATATTACAAGCCCGGAGAAATTGAAACGAAAATGCTGGCTTTCCAAGATGCCGGTAACCCCCCTCTGTACGTAATAGTTGAATGGAAGTACGAAGCCTCATTATTCAACCCAATGACGTGGAGATTAATAAAATCCCCAAGTATTTTCATCGAATATATAAAACTTTCGTCGATCGAATACAATACTGAAATAACGGTATGCCCAAAAATGAAAAAACCTGTCGTCAACAATCTGCCAACGGTCATGAAAACGAAATACTGCAAGTTCTAA
Protein
MMYNRTSATLMLQTIMLLMKKQNSTDVTNFPAGFSDDMELERCYDIYGCFSKAYPWTEHRPDNYFPASVESMAIRYPTFTRRNRKIPVLLHADNTDRIQNANLDANGPFYFISHGFLEGGHKKWIENLANALLDIDGNESATVIIVDWRGGSQPPYGQSVANIRLVGVMTAHLIHNIYEVLQLPHIDNFHFIGHSLGAHLGGYCGHELQKKFNLKLGRITGLDPAAPYFSRTVTLVRLDRSDAKYVDIIHSNAMPLYFSGFGISEPIGHVDFYPNGGSTQPGCKQSDSPNQSASYQQVVKYVGCNHERSHEYFTESIAPTCPFMAIQCESHEAFLAGNCTNCDNKHFCIPMGYHSYDLYKHLQIGGLVDTNTNIALYSLTGNAKPYCKVHYKVVLKVANSTESKNHGPESGKVTITLVDRNNNKTDRRSLNPEQKYYKPGEIETKMLAFQDAGNPPLYVIVEWKYEASLFNPMTWRLIKSPSIFIEYIKLSSIEYNTEITVCPKMKKPVVNNLPTVMKTKYCKF
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
A0A2W1BVF3
A0A212EQW0
A0A194Q337
A0A194R6H1
S4PA26
A0A2A4IZL5
+ More
A0A1E1WDL4 U5ETF1 A0A034VII1 A0A1Q3FPZ2 A0A1Q3FQ63 A0A1Q3FQ81 A0A1Q3FQ04 A0A0A1WQG5 B0XIK3 A0A0K8WL15 W8BBW7 A0A182GWT8 A0A1S4G2M0 Q16G20 A0A336LTX6 A0A2M4BLE1 A0A2M4BJS5 A0A2M4BJM1 D6X325 B4MTD0 T1PD82 A0A182XXZ0 A0A2M3Z3N1 A0A2M3Z3M1 Q16EM6 A0A182MH76 A0A1W4WBR7 A0A084VN29 A0A182QD72 A0A182SY61 Q29H54 B3NT00 B4H0A4 A0A0M3QZ73 A0A2M4AF95 A0A2M4AE66 B0XIK4 A0A3B0JWM8 B4Q1F5 B4MEL0 A0A1A9X5G7 A0A1W4WM28 W5JLI1 A0A1Q3FL03 A0A1I8NT75 B4R5B4 Q9W448 A0A2C9GS38 A0A182LD24 Q7QEY5 A0A182IM14 A0A182RMS1 B4I0G4 A0A1Y1LZ02 A0A1J1J590 A0A182W5C5 A0A182UNA0 A0A1Y1M085 B4L1H7 A0A182FJ93 B3MY91 B4JM78 A0A0L0C7N9 U4U8E2 A0A182NVG4 A0A182K4N6 A0A182UI44 A0A182XA52 A0A182P3A0 A0A026WVQ6 A0A0K8UW24 A0A232F843 A0A2A3E0I6 A0A1Y1M694 A0A1Y1M0D8 A0A1A9ZA71 A0A0K8VKT3 X2JDV7 A0A067QS20 A0A2M4BGM1 A0A182V7T6 A0A182JHM8 Q7QEY4 A0A2C9GS26 A0A2J7RLQ9 A0A1B0FEN7 A0A084VN28 E1ZYD8 T1GPL5 A0A182QJ84 A0A0C9PRB2
A0A1E1WDL4 U5ETF1 A0A034VII1 A0A1Q3FPZ2 A0A1Q3FQ63 A0A1Q3FQ81 A0A1Q3FQ04 A0A0A1WQG5 B0XIK3 A0A0K8WL15 W8BBW7 A0A182GWT8 A0A1S4G2M0 Q16G20 A0A336LTX6 A0A2M4BLE1 A0A2M4BJS5 A0A2M4BJM1 D6X325 B4MTD0 T1PD82 A0A182XXZ0 A0A2M3Z3N1 A0A2M3Z3M1 Q16EM6 A0A182MH76 A0A1W4WBR7 A0A084VN29 A0A182QD72 A0A182SY61 Q29H54 B3NT00 B4H0A4 A0A0M3QZ73 A0A2M4AF95 A0A2M4AE66 B0XIK4 A0A3B0JWM8 B4Q1F5 B4MEL0 A0A1A9X5G7 A0A1W4WM28 W5JLI1 A0A1Q3FL03 A0A1I8NT75 B4R5B4 Q9W448 A0A2C9GS38 A0A182LD24 Q7QEY5 A0A182IM14 A0A182RMS1 B4I0G4 A0A1Y1LZ02 A0A1J1J590 A0A182W5C5 A0A182UNA0 A0A1Y1M085 B4L1H7 A0A182FJ93 B3MY91 B4JM78 A0A0L0C7N9 U4U8E2 A0A182NVG4 A0A182K4N6 A0A182UI44 A0A182XA52 A0A182P3A0 A0A026WVQ6 A0A0K8UW24 A0A232F843 A0A2A3E0I6 A0A1Y1M694 A0A1Y1M0D8 A0A1A9ZA71 A0A0K8VKT3 X2JDV7 A0A067QS20 A0A2M4BGM1 A0A182V7T6 A0A182JHM8 Q7QEY4 A0A2C9GS26 A0A2J7RLQ9 A0A1B0FEN7 A0A084VN28 E1ZYD8 T1GPL5 A0A182QJ84 A0A0C9PRB2
Pubmed
28756777
22118469
26354079
23622113
25348373
25830018
+ More
24495485 26483478 17510324 18362917 19820115 17994087 25315136 25244985 24438588 15632085 17550304 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20966253 12364791 14747013 17210077 28004739 26108605 23537049 24508170 30249741 28648823 24845553 20798317
24495485 26483478 17510324 18362917 19820115 17994087 25315136 25244985 24438588 15632085 17550304 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20966253 12364791 14747013 17210077 28004739 26108605 23537049 24508170 30249741 28648823 24845553 20798317
EMBL
KZ149892
PZC78959.1
AGBW02013205
OWR43867.1
KQ459586
KPI97820.1
+ More
KQ460883 KPJ11461.1 GAIX01003444 JAA89116.1 NWSH01004745 PCG64714.1 GDQN01006005 JAT85049.1 GANO01002810 JAB57061.1 GAKP01017015 GAKP01017014 JAC41937.1 GFDL01005374 JAV29671.1 GFDL01005338 JAV29707.1 GFDL01005397 JAV29648.1 GFDL01005370 JAV29675.1 GBXI01013185 JAD01107.1 DS233324 EDS29427.1 GDHF01022836 GDHF01019022 GDHF01017568 GDHF01008176 GDHF01000456 JAI29478.1 JAI33292.1 JAI34746.1 JAI44138.1 JAI51858.1 GAMC01019201 JAB87354.1 JXUM01094109 KQ564104 KXJ72792.1 CH478344 EAT33186.1 UFQT01000107 SSX20089.1 GGFJ01004683 MBW53824.1 GGFJ01004092 MBW53233.1 GGFJ01004091 MBW53232.1 KQ971372 EFA10312.1 CH963851 EDW75369.1 KA646639 KA647024 AFP61268.1 GGFM01002369 MBW23120.1 GGFM01002361 MBW23112.1 CH478650 EAT32681.1 AXCM01001262 ATLV01014674 KE524978 KFB39373.1 AXCN02002018 CH379064 EAL31904.1 CH954180 EDV45969.1 CH479200 EDW29699.1 CP012528 ALC48879.1 GGFK01006135 MBW39456.1 GGFK01005681 MBW39002.1 EDS29428.1 OUUW01000003 SPP77756.1 CM000162 EDX02444.1 CH940664 EDW62985.1 ADMH02000812 ETN64966.1 GFDL01006755 JAV28290.1 CM000366 EDX17210.1 AE014298 AY058727 AAF46110.1 AAL13956.1 APCN01001149 AAAB01008846 EAA06347.4 CH480819 EDW52995.1 GEZM01043777 JAV78812.1 CVRI01000072 CRL07568.1 GEZM01043798 JAV78771.1 CH933810 EDW06698.1 CH902632 EDV32585.1 CH916371 EDV91839.1 JRES01000777 KNC28398.1 KB632220 ERL90189.1 KK107109 QOIP01000008 EZA59184.1 RLU19290.1 GDHF01021437 JAI30877.1 NNAY01000762 OXU26638.1 KZ288481 PBC25250.1 GEZM01043779 GEZM01043760 JAV78807.1 GEZM01043773 GEZM01043768 JAV78831.1 GDHF01012846 JAI39468.1 AHN59393.1 KK853376 KDR08050.1 GGFJ01003046 MBW52187.1 EAA06605.4 NEVH01002684 PNF41739.1 CCAG010003651 KFB39372.1 GL435204 EFN73837.1 CAQQ02390174 GBYB01003778 JAG73545.1
KQ460883 KPJ11461.1 GAIX01003444 JAA89116.1 NWSH01004745 PCG64714.1 GDQN01006005 JAT85049.1 GANO01002810 JAB57061.1 GAKP01017015 GAKP01017014 JAC41937.1 GFDL01005374 JAV29671.1 GFDL01005338 JAV29707.1 GFDL01005397 JAV29648.1 GFDL01005370 JAV29675.1 GBXI01013185 JAD01107.1 DS233324 EDS29427.1 GDHF01022836 GDHF01019022 GDHF01017568 GDHF01008176 GDHF01000456 JAI29478.1 JAI33292.1 JAI34746.1 JAI44138.1 JAI51858.1 GAMC01019201 JAB87354.1 JXUM01094109 KQ564104 KXJ72792.1 CH478344 EAT33186.1 UFQT01000107 SSX20089.1 GGFJ01004683 MBW53824.1 GGFJ01004092 MBW53233.1 GGFJ01004091 MBW53232.1 KQ971372 EFA10312.1 CH963851 EDW75369.1 KA646639 KA647024 AFP61268.1 GGFM01002369 MBW23120.1 GGFM01002361 MBW23112.1 CH478650 EAT32681.1 AXCM01001262 ATLV01014674 KE524978 KFB39373.1 AXCN02002018 CH379064 EAL31904.1 CH954180 EDV45969.1 CH479200 EDW29699.1 CP012528 ALC48879.1 GGFK01006135 MBW39456.1 GGFK01005681 MBW39002.1 EDS29428.1 OUUW01000003 SPP77756.1 CM000162 EDX02444.1 CH940664 EDW62985.1 ADMH02000812 ETN64966.1 GFDL01006755 JAV28290.1 CM000366 EDX17210.1 AE014298 AY058727 AAF46110.1 AAL13956.1 APCN01001149 AAAB01008846 EAA06347.4 CH480819 EDW52995.1 GEZM01043777 JAV78812.1 CVRI01000072 CRL07568.1 GEZM01043798 JAV78771.1 CH933810 EDW06698.1 CH902632 EDV32585.1 CH916371 EDV91839.1 JRES01000777 KNC28398.1 KB632220 ERL90189.1 KK107109 QOIP01000008 EZA59184.1 RLU19290.1 GDHF01021437 JAI30877.1 NNAY01000762 OXU26638.1 KZ288481 PBC25250.1 GEZM01043779 GEZM01043760 JAV78807.1 GEZM01043773 GEZM01043768 JAV78831.1 GDHF01012846 JAI39468.1 AHN59393.1 KK853376 KDR08050.1 GGFJ01003046 MBW52187.1 EAA06605.4 NEVH01002684 PNF41739.1 CCAG010003651 KFB39372.1 GL435204 EFN73837.1 CAQQ02390174 GBYB01003778 JAG73545.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000218220
UP000002320
UP000069940
+ More
UP000249989 UP000008820 UP000007266 UP000007798 UP000095301 UP000076408 UP000075883 UP000192221 UP000030765 UP000075886 UP000075901 UP000001819 UP000008711 UP000008744 UP000092553 UP000268350 UP000002282 UP000008792 UP000091820 UP000192223 UP000000673 UP000095300 UP000000304 UP000000803 UP000075840 UP000075882 UP000007062 UP000075880 UP000075900 UP000001292 UP000183832 UP000075920 UP000075903 UP000009192 UP000069272 UP000007801 UP000001070 UP000037069 UP000030742 UP000075884 UP000075881 UP000075902 UP000076407 UP000075885 UP000053097 UP000279307 UP000215335 UP000242457 UP000092445 UP000027135 UP000235965 UP000092444 UP000000311 UP000015102
UP000249989 UP000008820 UP000007266 UP000007798 UP000095301 UP000076408 UP000075883 UP000192221 UP000030765 UP000075886 UP000075901 UP000001819 UP000008711 UP000008744 UP000092553 UP000268350 UP000002282 UP000008792 UP000091820 UP000192223 UP000000673 UP000095300 UP000000304 UP000000803 UP000075840 UP000075882 UP000007062 UP000075880 UP000075900 UP000001292 UP000183832 UP000075920 UP000075903 UP000009192 UP000069272 UP000007801 UP000001070 UP000037069 UP000030742 UP000075884 UP000075881 UP000075902 UP000076407 UP000075885 UP000053097 UP000279307 UP000215335 UP000242457 UP000092445 UP000027135 UP000235965 UP000092444 UP000000311 UP000015102
Pfam
PF00151 Lipase
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BVF3
A0A212EQW0
A0A194Q337
A0A194R6H1
S4PA26
A0A2A4IZL5
+ More
A0A1E1WDL4 U5ETF1 A0A034VII1 A0A1Q3FPZ2 A0A1Q3FQ63 A0A1Q3FQ81 A0A1Q3FQ04 A0A0A1WQG5 B0XIK3 A0A0K8WL15 W8BBW7 A0A182GWT8 A0A1S4G2M0 Q16G20 A0A336LTX6 A0A2M4BLE1 A0A2M4BJS5 A0A2M4BJM1 D6X325 B4MTD0 T1PD82 A0A182XXZ0 A0A2M3Z3N1 A0A2M3Z3M1 Q16EM6 A0A182MH76 A0A1W4WBR7 A0A084VN29 A0A182QD72 A0A182SY61 Q29H54 B3NT00 B4H0A4 A0A0M3QZ73 A0A2M4AF95 A0A2M4AE66 B0XIK4 A0A3B0JWM8 B4Q1F5 B4MEL0 A0A1A9X5G7 A0A1W4WM28 W5JLI1 A0A1Q3FL03 A0A1I8NT75 B4R5B4 Q9W448 A0A2C9GS38 A0A182LD24 Q7QEY5 A0A182IM14 A0A182RMS1 B4I0G4 A0A1Y1LZ02 A0A1J1J590 A0A182W5C5 A0A182UNA0 A0A1Y1M085 B4L1H7 A0A182FJ93 B3MY91 B4JM78 A0A0L0C7N9 U4U8E2 A0A182NVG4 A0A182K4N6 A0A182UI44 A0A182XA52 A0A182P3A0 A0A026WVQ6 A0A0K8UW24 A0A232F843 A0A2A3E0I6 A0A1Y1M694 A0A1Y1M0D8 A0A1A9ZA71 A0A0K8VKT3 X2JDV7 A0A067QS20 A0A2M4BGM1 A0A182V7T6 A0A182JHM8 Q7QEY4 A0A2C9GS26 A0A2J7RLQ9 A0A1B0FEN7 A0A084VN28 E1ZYD8 T1GPL5 A0A182QJ84 A0A0C9PRB2
A0A1E1WDL4 U5ETF1 A0A034VII1 A0A1Q3FPZ2 A0A1Q3FQ63 A0A1Q3FQ81 A0A1Q3FQ04 A0A0A1WQG5 B0XIK3 A0A0K8WL15 W8BBW7 A0A182GWT8 A0A1S4G2M0 Q16G20 A0A336LTX6 A0A2M4BLE1 A0A2M4BJS5 A0A2M4BJM1 D6X325 B4MTD0 T1PD82 A0A182XXZ0 A0A2M3Z3N1 A0A2M3Z3M1 Q16EM6 A0A182MH76 A0A1W4WBR7 A0A084VN29 A0A182QD72 A0A182SY61 Q29H54 B3NT00 B4H0A4 A0A0M3QZ73 A0A2M4AF95 A0A2M4AE66 B0XIK4 A0A3B0JWM8 B4Q1F5 B4MEL0 A0A1A9X5G7 A0A1W4WM28 W5JLI1 A0A1Q3FL03 A0A1I8NT75 B4R5B4 Q9W448 A0A2C9GS38 A0A182LD24 Q7QEY5 A0A182IM14 A0A182RMS1 B4I0G4 A0A1Y1LZ02 A0A1J1J590 A0A182W5C5 A0A182UNA0 A0A1Y1M085 B4L1H7 A0A182FJ93 B3MY91 B4JM78 A0A0L0C7N9 U4U8E2 A0A182NVG4 A0A182K4N6 A0A182UI44 A0A182XA52 A0A182P3A0 A0A026WVQ6 A0A0K8UW24 A0A232F843 A0A2A3E0I6 A0A1Y1M694 A0A1Y1M0D8 A0A1A9ZA71 A0A0K8VKT3 X2JDV7 A0A067QS20 A0A2M4BGM1 A0A182V7T6 A0A182JHM8 Q7QEY4 A0A2C9GS26 A0A2J7RLQ9 A0A1B0FEN7 A0A084VN28 E1ZYD8 T1GPL5 A0A182QJ84 A0A0C9PRB2
PDB
1W52
E-value=1.56179e-60,
Score=591
Ontologies
PATHWAY
GO
PANTHER
Topology
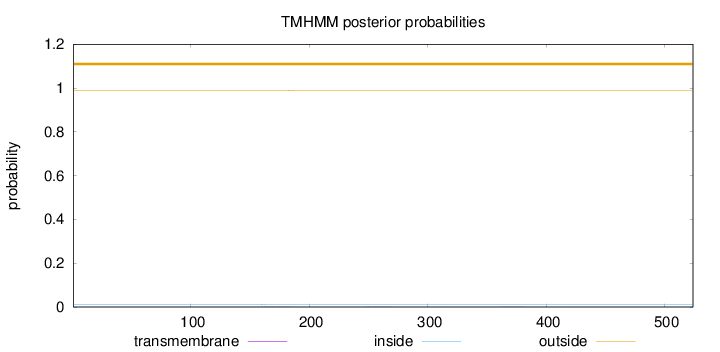
Subcellular location
Secreted
Length:
524
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03371
Exp number, first 60 AAs:
0.00127
Total prob of N-in:
0.01137
outside
1 - 524
Population Genetic Test Statistics
Pi
238.55677
Theta
168.491456
Tajima's D
0.822985
CLR
0.289115
CSRT
0.611019449027549
Interpretation
Uncertain
