Gene
KWMTBOMO13046
Pre Gene Modal
BGIBMGA000293
Annotation
PREDICTED:_GTPase_Era?_mitochondrial_[Papilio_xuthus]
Location in the cell
Mitochondrial Reliability : 1.395 Nuclear Reliability : 2.091
Sequence
CDS
ATGCTCCCTCGTCGTATCGTTAATTATTCATTTCAACCACAACATGCTGAAAATAAAAACATAGGTAAAATCGTACATGTTGCGATTATTGGAGCCCCAAATTCAGGAAAAAGTACTTTAATAAACAAAATAATTGACCGAAAGATATGTGCTGCTTCAAATAAAGTTCATACAACTACCAAATTAGTAAGAGCAATGTGTTACTGCAATGACACACAGATTGTATTTCTGGATACACCGGGTGTTGTAACTGATAGAGAACAAAAGAAATACAACCTCCCACCTTCAATGTTAGGATCATGTCACAAAAGTCTCAGATGTGCAGATGTAGTTGGTGTTGTCCATGATGCATCATGTAAATATATCAGAGAATCTTTACACAAGGATGTTGTTGAAATGCTGAACTCTGTACAAGATATGCCAAGTTTTCTTATCATAAATAAGGTTGATAAGTTAAGGTCAAAGAAGCAGTTATTGACTTTAGTGAGGAATCTCACTAATGGATTTATTGCTGGAAACTCTATTCCTGGTCCCACAAATCCTAATAAACAGGAGAGAGGCTTCAGTCATTTTTCGGATGTGTTTATGGTATCTGCTTTGAATGGTGATGGAGTTTCAGATATAAGGGAGTACCTTGTCAACAACGCCAAGCCCGGAAAGTTTCATTATTCAACTGAAGAATGGACTGACCAAACGCCGAAATCAGTCATAGAAGACGCTGTGAGAGCGAAATTCTTAGATTTTTTGCATCAAGAAATACCATACAATTTGAAAGTGAAATTAGATTATTACGAAGAGATCGATAACGAAGATAAAATCATATGTTCAGTATCTGTAGAGTGTCCATCAGAAAGGCTGATGCGACTAATAAGCGGCGCCGGCGGCGGCCGCCTCCAACAGATAAAGTCTTCTGTTCGGAATGATTTGATAGACGTCTTCAGGAAAACGGTGATCTTAGACTTGCAATTGCACGTGAAAAATAATTTTTTGTTAGCTAGTTAG
Protein
MLPRRIVNYSFQPQHAENKNIGKIVHVAIIGAPNSGKSTLINKIIDRKICAASNKVHTTTKLVRAMCYCNDTQIVFLDTPGVVTDREQKKYNLPPSMLGSCHKSLRCADVVGVVHDASCKYIRESLHKDVVEMLNSVQDMPSFLIINKVDKLRSKKQLLTLVRNLTNGFIAGNSIPGPTNPNKQERGFSHFSDVFMVSALNGDGVSDIREYLVNNAKPGKFHYSTEEWTDQTPKSVIEDAVRAKFLDFLHQEIPYNLKVKLDYYEEIDNEDKIICSVSVECPSERLMRLISGAGGGRLQQIKSSVRNDLIDVFRKTVILDLQLHVKNNFLLAS
Summary
Similarity
Belongs to the helicase family. Dicer subfamily.
Uniprot
H9ISR6
A0A2H1VCV0
A0A2A4ITF8
S4PL44
A0A2W1BXI4
A0A212FLX9
+ More
A0A194QWP5 A0A3S2NRC3 A0A194PWW9 A0A0L7LKY2 D6WW14 B3MJ76 J3JX59 B4MR53 A0A1L8DWN5 Q9VG07 Q179N6 B4HGD0 A0A1L8DWN0 B4R1Y3 N6T0R5 A0A1S4FB92 A0A034WP93 B4KLZ8 B4PPU8 A0A1W4UKF9 A0A182G5E5 A0A3B0J750 B0WML6 B4J4A9 B4LIJ6 W8CEG1 A0A1Q3F6L3 A0A0M4EIS5 B4GBU0 B5E129 A0A0K8WKR8 A0A2M4BQN8 A0A2M4BQY0 A0A1Y1LVM3 A0A1B0CFP8 A0A1A9YBF1 A0A2M3ZEJ9 A0A182QTP0 A0A1A9WDV7 A0A2M4BQN0 A0A2M4BQT9 A0A0L0C9H6 B3P1A1 A0A0A1XNY4 A0A182FUD8 W5JJK8 A0A1B0D336 Q7QG96 A0A182NPH9 A0A182KCP2 A0A1B0ACW8 A0A2R7WEM6 A0A182X0T9 A0A182KY32 A0A182HFL3 A0A182YA76 A0A182V9T9 A0A182PBX8 A0A0A9WH67 A0A182S6X3 A0A182UBA3 A0A084WLQ8 A0A182M5T0 A0A182W2F3 A0A182RR02 A0A1B0B2F4 A0A1I8PGV0 A0A1I8N128 A0A1J1J7M4 A0A1A9UVA5 A0A1B6CJJ9 A0A2S2Q172 A0A069DRU1 A0A0J7L5F6 A0A0C9RKM2 A0A1B0FE40 A0A232F592 A0A088APV4 A0A1B6EQ39 K7IXC9 A0A2R5LJ48 E9IVM6 A0A293LUT1 A0A026WDK5 E2A153 A0A0V0GCD9 A0A195CSS7 A0A2A3EJS7 A0A2H8TN51 A0A195DZK3 A0A131XZE4 A0A0P4VMW1 A0A0K8R7F7 A0A195FLI1
A0A194QWP5 A0A3S2NRC3 A0A194PWW9 A0A0L7LKY2 D6WW14 B3MJ76 J3JX59 B4MR53 A0A1L8DWN5 Q9VG07 Q179N6 B4HGD0 A0A1L8DWN0 B4R1Y3 N6T0R5 A0A1S4FB92 A0A034WP93 B4KLZ8 B4PPU8 A0A1W4UKF9 A0A182G5E5 A0A3B0J750 B0WML6 B4J4A9 B4LIJ6 W8CEG1 A0A1Q3F6L3 A0A0M4EIS5 B4GBU0 B5E129 A0A0K8WKR8 A0A2M4BQN8 A0A2M4BQY0 A0A1Y1LVM3 A0A1B0CFP8 A0A1A9YBF1 A0A2M3ZEJ9 A0A182QTP0 A0A1A9WDV7 A0A2M4BQN0 A0A2M4BQT9 A0A0L0C9H6 B3P1A1 A0A0A1XNY4 A0A182FUD8 W5JJK8 A0A1B0D336 Q7QG96 A0A182NPH9 A0A182KCP2 A0A1B0ACW8 A0A2R7WEM6 A0A182X0T9 A0A182KY32 A0A182HFL3 A0A182YA76 A0A182V9T9 A0A182PBX8 A0A0A9WH67 A0A182S6X3 A0A182UBA3 A0A084WLQ8 A0A182M5T0 A0A182W2F3 A0A182RR02 A0A1B0B2F4 A0A1I8PGV0 A0A1I8N128 A0A1J1J7M4 A0A1A9UVA5 A0A1B6CJJ9 A0A2S2Q172 A0A069DRU1 A0A0J7L5F6 A0A0C9RKM2 A0A1B0FE40 A0A232F592 A0A088APV4 A0A1B6EQ39 K7IXC9 A0A2R5LJ48 E9IVM6 A0A293LUT1 A0A026WDK5 E2A153 A0A0V0GCD9 A0A195CSS7 A0A2A3EJS7 A0A2H8TN51 A0A195DZK3 A0A131XZE4 A0A0P4VMW1 A0A0K8R7F7 A0A195FLI1
Pubmed
19121390
23622113
28756777
22118469
26354079
26227816
+ More
18362917 19820115 17994087 22516182 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17510324 23537049 25348373 17550304 26483478 18057021 24495485 15632085 28004739 26108605 25830018 20920257 23761445 12364791 20966253 25244985 25401762 26823975 24438588 25315136 26334808 28648823 20075255 21282665 24508170 30249741 20798317 27129103
18362917 19820115 17994087 22516182 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17510324 23537049 25348373 17550304 26483478 18057021 24495485 15632085 28004739 26108605 25830018 20920257 23761445 12364791 20966253 25244985 25401762 26823975 24438588 25315136 26334808 28648823 20075255 21282665 24508170 30249741 20798317 27129103
EMBL
BABH01013291
ODYU01001851
SOQ38631.1
NWSH01007301
PCG63011.1
GAIX01004230
+ More
JAA88330.1 KZ149892 PZC79031.1 AGBW02007691 OWR54742.1 KQ461073 KPJ09729.1 RSAL01000005 RVE54362.1 KQ459586 KPI97817.1 JTDY01000747 KOB76024.1 KQ971361 EFA08204.1 CH902619 EDV38170.1 BT127827 AEE62789.1 CH963849 EDW74592.1 GFDF01003251 JAV10833.1 AE014297 BT031201 AAF54886.2 ABY20442.1 CH477348 EAT42922.1 CH480815 EDW42378.1 GFDF01003252 JAV10832.1 CM000364 EDX13135.1 APGK01056553 APGK01056554 KB741277 KB631816 ENN71073.1 ERL86518.1 GAKP01002815 JAC56137.1 CH933808 EDW10787.1 CM000160 EDW97175.1 JXUM01007080 KQ560230 KXJ83719.1 OUUW01000001 SPP75933.1 DS231998 EDS31087.1 CH916367 EDW00589.1 CH940648 EDW60366.1 KRF79384.1 GAMC01000256 JAC06300.1 GFDL01011892 JAV23153.1 CP012524 ALC41055.1 CH479181 EDW31319.1 CM000071 EDY68686.1 GDHF01000622 JAI51692.1 GGFJ01006255 MBW55396.1 GGFJ01006242 MBW55383.1 GEZM01045908 GEZM01045907 JAV77629.1 AJWK01010250 GGFM01006225 MBW26976.1 AXCN02001829 GGFJ01006254 MBW55395.1 GGFJ01006241 MBW55382.1 JRES01000697 KNC29093.1 CH954181 EDV49290.1 GBXI01001586 JAD12706.1 ADMH02000962 ETN64562.1 AJVK01023227 AAAB01008838 EAA05838.3 KK854700 PTY18008.1 APCN01006364 GBHO01044611 GBHO01037740 GBHO01037738 GBHO01037736 GBHO01037733 GBHO01023638 GBHO01005792 GBRD01010349 GBRD01007685 GBRD01007684 GDHC01002183 JAF98992.1 JAG05864.1 JAG05866.1 JAG05868.1 JAG05871.1 JAG19966.1 JAG37812.1 JAG55475.1 JAQ16446.1 ATLV01024269 KE525351 KFB51152.1 AXCM01001761 JXJN01007622 JXJN01007623 CVRI01000075 CRL08391.1 GEDC01023681 JAS13617.1 GGMS01002303 MBY71506.1 GBGD01002229 JAC86660.1 LBMM01000509 KMQ98187.1 GBYB01013812 GBYB01013813 JAG83579.1 JAG83580.1 CCAG010018330 NNAY01000980 OXU25633.1 GECZ01029712 JAS40057.1 AAZX01000018 GGLE01005342 MBY09468.1 GL766315 EFZ15377.1 GFWV01006851 MAA31581.1 KK107260 QOIP01000001 EZA54170.1 RLU27653.1 GL435707 EFN72820.1 GECL01001095 JAP05029.1 KQ977381 KYN03169.1 KZ288232 PBC31512.1 GFXV01003546 MBW15351.1 KQ979999 KYN18303.1 GEFM01003193 JAP72603.1 GDKW01002781 JAI53814.1 GADI01007394 JAA66414.1 KQ981490 KYN41273.1
JAA88330.1 KZ149892 PZC79031.1 AGBW02007691 OWR54742.1 KQ461073 KPJ09729.1 RSAL01000005 RVE54362.1 KQ459586 KPI97817.1 JTDY01000747 KOB76024.1 KQ971361 EFA08204.1 CH902619 EDV38170.1 BT127827 AEE62789.1 CH963849 EDW74592.1 GFDF01003251 JAV10833.1 AE014297 BT031201 AAF54886.2 ABY20442.1 CH477348 EAT42922.1 CH480815 EDW42378.1 GFDF01003252 JAV10832.1 CM000364 EDX13135.1 APGK01056553 APGK01056554 KB741277 KB631816 ENN71073.1 ERL86518.1 GAKP01002815 JAC56137.1 CH933808 EDW10787.1 CM000160 EDW97175.1 JXUM01007080 KQ560230 KXJ83719.1 OUUW01000001 SPP75933.1 DS231998 EDS31087.1 CH916367 EDW00589.1 CH940648 EDW60366.1 KRF79384.1 GAMC01000256 JAC06300.1 GFDL01011892 JAV23153.1 CP012524 ALC41055.1 CH479181 EDW31319.1 CM000071 EDY68686.1 GDHF01000622 JAI51692.1 GGFJ01006255 MBW55396.1 GGFJ01006242 MBW55383.1 GEZM01045908 GEZM01045907 JAV77629.1 AJWK01010250 GGFM01006225 MBW26976.1 AXCN02001829 GGFJ01006254 MBW55395.1 GGFJ01006241 MBW55382.1 JRES01000697 KNC29093.1 CH954181 EDV49290.1 GBXI01001586 JAD12706.1 ADMH02000962 ETN64562.1 AJVK01023227 AAAB01008838 EAA05838.3 KK854700 PTY18008.1 APCN01006364 GBHO01044611 GBHO01037740 GBHO01037738 GBHO01037736 GBHO01037733 GBHO01023638 GBHO01005792 GBRD01010349 GBRD01007685 GBRD01007684 GDHC01002183 JAF98992.1 JAG05864.1 JAG05866.1 JAG05868.1 JAG05871.1 JAG19966.1 JAG37812.1 JAG55475.1 JAQ16446.1 ATLV01024269 KE525351 KFB51152.1 AXCM01001761 JXJN01007622 JXJN01007623 CVRI01000075 CRL08391.1 GEDC01023681 JAS13617.1 GGMS01002303 MBY71506.1 GBGD01002229 JAC86660.1 LBMM01000509 KMQ98187.1 GBYB01013812 GBYB01013813 JAG83579.1 JAG83580.1 CCAG010018330 NNAY01000980 OXU25633.1 GECZ01029712 JAS40057.1 AAZX01000018 GGLE01005342 MBY09468.1 GL766315 EFZ15377.1 GFWV01006851 MAA31581.1 KK107260 QOIP01000001 EZA54170.1 RLU27653.1 GL435707 EFN72820.1 GECL01001095 JAP05029.1 KQ977381 KYN03169.1 KZ288232 PBC31512.1 GFXV01003546 MBW15351.1 KQ979999 KYN18303.1 GEFM01003193 JAP72603.1 GDKW01002781 JAI53814.1 GADI01007394 JAA66414.1 KQ981490 KYN41273.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000283053
UP000053268
+ More
UP000037510 UP000007266 UP000007801 UP000007798 UP000000803 UP000008820 UP000001292 UP000000304 UP000019118 UP000030742 UP000009192 UP000002282 UP000192221 UP000069940 UP000249989 UP000268350 UP000002320 UP000001070 UP000008792 UP000092553 UP000008744 UP000001819 UP000092461 UP000092443 UP000075886 UP000091820 UP000037069 UP000008711 UP000069272 UP000000673 UP000092462 UP000007062 UP000075884 UP000075881 UP000092445 UP000076407 UP000075882 UP000075840 UP000076408 UP000075903 UP000075885 UP000075901 UP000075902 UP000030765 UP000075883 UP000075920 UP000075900 UP000092460 UP000095300 UP000095301 UP000183832 UP000078200 UP000036403 UP000092444 UP000215335 UP000005203 UP000002358 UP000053097 UP000279307 UP000000311 UP000078542 UP000242457 UP000078492 UP000078541
UP000037510 UP000007266 UP000007801 UP000007798 UP000000803 UP000008820 UP000001292 UP000000304 UP000019118 UP000030742 UP000009192 UP000002282 UP000192221 UP000069940 UP000249989 UP000268350 UP000002320 UP000001070 UP000008792 UP000092553 UP000008744 UP000001819 UP000092461 UP000092443 UP000075886 UP000091820 UP000037069 UP000008711 UP000069272 UP000000673 UP000092462 UP000007062 UP000075884 UP000075881 UP000092445 UP000076407 UP000075882 UP000075840 UP000076408 UP000075903 UP000075885 UP000075901 UP000075902 UP000030765 UP000075883 UP000075920 UP000075900 UP000092460 UP000095300 UP000095301 UP000183832 UP000078200 UP000036403 UP000092444 UP000215335 UP000005203 UP000002358 UP000053097 UP000279307 UP000000311 UP000078542 UP000242457 UP000078492 UP000078541
Pfam
Interpro
IPR027417
P-loop_NTPase
+ More
IPR009019 KH_sf_prok-type
IPR015946 KH_dom-like_a/b
IPR030388 G_ERA_dom
IPR005225 Small_GTP-bd_dom
IPR005662 GTP-bd_Era
IPR006073 GTP_binding_domain
IPR003100 PAZ_dom
IPR006935 Helicase/UvrB_N
IPR000999 RNase_III_dom
IPR038248 Dicer_dimer_sf
IPR005034 Dicer_dimerisation_dom
IPR036389 RNase_III_sf
IPR014720 dsRBD_dom
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR013005 Ribosomal_uL4/L1e
IPR002136 Ribosomal_L4/L1e
IPR023574 Ribosomal_L4_dom_sf
IPR004044 KH_dom_type_2
IPR009019 KH_sf_prok-type
IPR015946 KH_dom-like_a/b
IPR030388 G_ERA_dom
IPR005225 Small_GTP-bd_dom
IPR005662 GTP-bd_Era
IPR006073 GTP_binding_domain
IPR003100 PAZ_dom
IPR006935 Helicase/UvrB_N
IPR000999 RNase_III_dom
IPR038248 Dicer_dimer_sf
IPR005034 Dicer_dimerisation_dom
IPR036389 RNase_III_sf
IPR014720 dsRBD_dom
IPR001650 Helicase_C
IPR014001 Helicase_ATP-bd
IPR013005 Ribosomal_uL4/L1e
IPR002136 Ribosomal_L4/L1e
IPR023574 Ribosomal_L4_dom_sf
IPR004044 KH_dom_type_2
ProteinModelPortal
H9ISR6
A0A2H1VCV0
A0A2A4ITF8
S4PL44
A0A2W1BXI4
A0A212FLX9
+ More
A0A194QWP5 A0A3S2NRC3 A0A194PWW9 A0A0L7LKY2 D6WW14 B3MJ76 J3JX59 B4MR53 A0A1L8DWN5 Q9VG07 Q179N6 B4HGD0 A0A1L8DWN0 B4R1Y3 N6T0R5 A0A1S4FB92 A0A034WP93 B4KLZ8 B4PPU8 A0A1W4UKF9 A0A182G5E5 A0A3B0J750 B0WML6 B4J4A9 B4LIJ6 W8CEG1 A0A1Q3F6L3 A0A0M4EIS5 B4GBU0 B5E129 A0A0K8WKR8 A0A2M4BQN8 A0A2M4BQY0 A0A1Y1LVM3 A0A1B0CFP8 A0A1A9YBF1 A0A2M3ZEJ9 A0A182QTP0 A0A1A9WDV7 A0A2M4BQN0 A0A2M4BQT9 A0A0L0C9H6 B3P1A1 A0A0A1XNY4 A0A182FUD8 W5JJK8 A0A1B0D336 Q7QG96 A0A182NPH9 A0A182KCP2 A0A1B0ACW8 A0A2R7WEM6 A0A182X0T9 A0A182KY32 A0A182HFL3 A0A182YA76 A0A182V9T9 A0A182PBX8 A0A0A9WH67 A0A182S6X3 A0A182UBA3 A0A084WLQ8 A0A182M5T0 A0A182W2F3 A0A182RR02 A0A1B0B2F4 A0A1I8PGV0 A0A1I8N128 A0A1J1J7M4 A0A1A9UVA5 A0A1B6CJJ9 A0A2S2Q172 A0A069DRU1 A0A0J7L5F6 A0A0C9RKM2 A0A1B0FE40 A0A232F592 A0A088APV4 A0A1B6EQ39 K7IXC9 A0A2R5LJ48 E9IVM6 A0A293LUT1 A0A026WDK5 E2A153 A0A0V0GCD9 A0A195CSS7 A0A2A3EJS7 A0A2H8TN51 A0A195DZK3 A0A131XZE4 A0A0P4VMW1 A0A0K8R7F7 A0A195FLI1
A0A194QWP5 A0A3S2NRC3 A0A194PWW9 A0A0L7LKY2 D6WW14 B3MJ76 J3JX59 B4MR53 A0A1L8DWN5 Q9VG07 Q179N6 B4HGD0 A0A1L8DWN0 B4R1Y3 N6T0R5 A0A1S4FB92 A0A034WP93 B4KLZ8 B4PPU8 A0A1W4UKF9 A0A182G5E5 A0A3B0J750 B0WML6 B4J4A9 B4LIJ6 W8CEG1 A0A1Q3F6L3 A0A0M4EIS5 B4GBU0 B5E129 A0A0K8WKR8 A0A2M4BQN8 A0A2M4BQY0 A0A1Y1LVM3 A0A1B0CFP8 A0A1A9YBF1 A0A2M3ZEJ9 A0A182QTP0 A0A1A9WDV7 A0A2M4BQN0 A0A2M4BQT9 A0A0L0C9H6 B3P1A1 A0A0A1XNY4 A0A182FUD8 W5JJK8 A0A1B0D336 Q7QG96 A0A182NPH9 A0A182KCP2 A0A1B0ACW8 A0A2R7WEM6 A0A182X0T9 A0A182KY32 A0A182HFL3 A0A182YA76 A0A182V9T9 A0A182PBX8 A0A0A9WH67 A0A182S6X3 A0A182UBA3 A0A084WLQ8 A0A182M5T0 A0A182W2F3 A0A182RR02 A0A1B0B2F4 A0A1I8PGV0 A0A1I8N128 A0A1J1J7M4 A0A1A9UVA5 A0A1B6CJJ9 A0A2S2Q172 A0A069DRU1 A0A0J7L5F6 A0A0C9RKM2 A0A1B0FE40 A0A232F592 A0A088APV4 A0A1B6EQ39 K7IXC9 A0A2R5LJ48 E9IVM6 A0A293LUT1 A0A026WDK5 E2A153 A0A0V0GCD9 A0A195CSS7 A0A2A3EJS7 A0A2H8TN51 A0A195DZK3 A0A131XZE4 A0A0P4VMW1 A0A0K8R7F7 A0A195FLI1
PDB
3IEV
E-value=3.48123e-20,
Score=241
Ontologies
GO
Topology
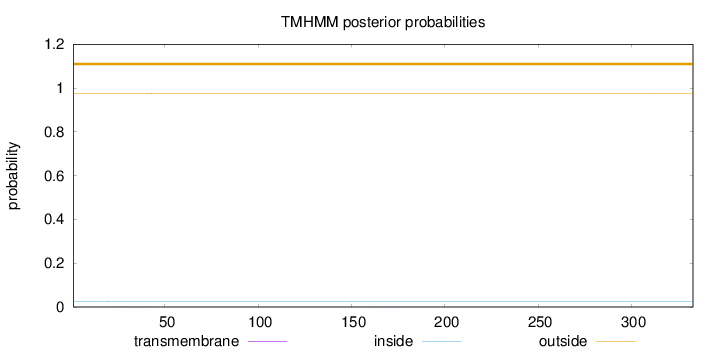
Length:
333
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01504
Exp number, first 60 AAs:
0.0091
Total prob of N-in:
0.02561
outside
1 - 333
Population Genetic Test Statistics
Pi
190.50993
Theta
150.629882
Tajima's D
-1.000536
CLR
190.704114
CSRT
0.138293085345733
Interpretation
Uncertain
