Gene
KWMTBOMO13041 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000292
Annotation
dihydrolipoamide_dehydrogenase_[Bombyx_mori]
Full name
Dihydrolipoyl dehydrogenase
+ More
Dihydrolipoyl dehydrogenase, mitochondrial
Dihydrolipoyl dehydrogenase, mitochondrial
Alternative Name
Dihydrolipoamide dehydrogenase
E3
E3
Location in the cell
Cytoplasmic Reliability : 1.783 Mitochondrial Reliability : 2.428
Sequence
CDS
ATGGGCTATAAATTTCTAAAACTTGCCTCTCCTACCTTTAGAAGTGGAAGTTTAGTACGAATTGCTACAAGACAATATGCTACCACACATGATGCAGATCTTGTTGTTATCGGTTCGGGCCCTGGTGGATACGTAGCAGCTATTAAAGCTGCCCAGCTTGGCATGAAGGTGGTCTCAGTAGAAAAGGACCCTACTTTAGGAGGTACTTGTCTCAATGTTGGATGTATACCATCAAAAGCTTTACTGCACAACTCACATCTTTACCATATGGCCAAACATGACTTCAAGCAAAGGGGTATTGAAACTGGTGAAGTTACATTTGACTTCAAAAAGATGATGGAATATAAAGCTAATGCTGTTAAAGGCCTCACAGGAGGTATTGCAATGCTCTTTCAGAAGAACAAGGTTAATCTTGTCAAAGGTGTAGGAACTATAGTTGCCCCAAATAAAGTTGAAGTACACGGAGAGAAGGGTGTTGAGACTGTTAATACCAAAAATATTCTAATCGCTTCTGGGTCTGAAGTTACACCTTTCCCTGGAGTTACTTTTGATGAGAAACAAATAATCACATCGACTGGAGCACTTTCCTTGGAGTCAGTACCGAAAAAAATGTTGGTGATTGGAGCTGGAGTTATCGGTTTGGAGCTTGGTTCTGTTTATCAAAGACTTGGTGCTGATGTTACTGCAATTGAATTCCTAACTAGCATTGGAGGTGTTGGTATTGATGGAGAGGTAGCCAAGACTCTTCAGAAAATCTTAAGCAAACAAGGAATGAAATTCAAACTGGGAACTAAAGTTTTAGGGGTTAAGAAAGAAGGTAGTACTATCAAAGTTGATGTTGAAGCTGCTAAAGGTGGCAATAAGGAAGTGTTGGATTGCGATGTGGTGCTAATTTCAATTGGTCGTCGCCCTTACACTAAAGGTCTCGGGCTCGACAAAGTGGGCATCGCTTTGGACGACCGCGGCCGGATTCCTGTCAACAACAAATTCCAAACTACTGTGCCCGGAATTTACGCTATTGGTGATGTAATTCACGGACCAATGTTGGCTCACAAAGCGGAGGACGAAGGTATCGTCTGCGTGGAAGGTATTAAGGGTATGCCCGTCCACTTCAACTACGATGCAATTCCGTCGGTTATTTACACTTCACCCGAAGTTGGCTGGGTCGGAAAGACTGAAGAAGATTTAAAGAAAGAAGGCAGAGCTTACAAAGTGGGTAAATTCCCGTTCCTGGCGAACTCCAGAGCGAAAACGAATGGGGAAACTGAAGGTTTCGTCAAAGTGCTTTCCGATAAGACTACAGACGTAATCCTTGGGACTCACATTATTGGCCCTGGCGGTGGTGAGTTGATCAACGAGGCTGTACTCGCGCAGGAGTATGGTGCCGCTGCTGAAGACGTTGCGAGAGTCTGTCACGCGCATCCTACATGTGCCGAAGCGCTACGTGAAGCAAATCTAGCAGCGTACTCCGGAAAACCAATCAACTTCTAA
Protein
MGYKFLKLASPTFRSGSLVRIATRQYATTHDADLVVIGSGPGGYVAAIKAAQLGMKVVSVEKDPTLGGTCLNVGCIPSKALLHNSHLYHMAKHDFKQRGIETGEVTFDFKKMMEYKANAVKGLTGGIAMLFQKNKVNLVKGVGTIVAPNKVEVHGEKGVETVNTKNILIASGSEVTPFPGVTFDEKQIITSTGALSLESVPKKMLVIGAGVIGLELGSVYQRLGADVTAIEFLTSIGGVGIDGEVAKTLQKILSKQGMKFKLGTKVLGVKKEGSTIKVDVEAAKGGNKEVLDCDVVLISIGRRPYTKGLGLDKVGIALDDRGRIPVNNKFQTTVPGIYAIGDVIHGPMLAHKAEDEGIVCVEGIKGMPVHFNYDAIPSVIYTSPEVGWVGKTEEDLKKEGRAYKVGKFPFLANSRAKTNGETEGFVKVLSDKTTDVILGTHIIGPGGGELINEAVLAQEYGAAAEDVARVCHAHPTCAEALREANLAAYSGKPINF
Summary
Catalytic Activity
(R)-N(6)-dihydrolipoyl-L-lysyl-[protein] + NAD(+) = (R)-N(6)-lipoyl-L-lysyl-[protein] + H(+) + NADH
Cofactor
FAD
Subunit
Homodimer.
Miscellaneous
The active site is a redox-active disulfide bond.
Similarity
Belongs to the class-I pyridine nucleotide-disulfide oxidoreductase family.
Keywords
Cytoplasm
Disulfide bond
FAD
Flavoprotein
Glycolysis
NAD
Oxidoreductase
Redox-active center
Alternative splicing
Complete proteome
Fatty acid biosynthesis
Fatty acid metabolism
Lipid biosynthesis
Lipid metabolism
Mitochondrion
Reference proteome
Transit peptide
Feature
chain Dihydrolipoyl dehydrogenase
splice variant In isoform b.
splice variant In isoform b.
Uniprot
Q8MUB0
D6R773
I4DMH5
S4P991
O18480
A0A3S2M9V9
+ More
A0A212FM25 A0A2H1VCT0 A0A2W1C0K5 A0A194Q332 I4DJF2 A0A2A4IX85 A0A0L0CMD7 B4L0F4 W8C1K5 T1PHL2 A0A1I8NPA9 A0A0K8TZ55 A0A034VK02 A0A0A1XNC7 B4LIF2 D3TRU9 A0A1A9URY6 A0A1L8EEI8 Q9VVL7 K7PQ56 B3M3U9 A0A1B0A6P9 A0A1L6K358 K7PR60 A0A0J9RYP2 B4HLB4 A0A1Y1L6U1 A0A1W4W542 K7PQS7 B4PJH9 A0A0M5JB65 K7PQU4 B4N4T3 A0A1L6K360 K7PQU5 K7PQ54 B4QPB7 A0A1B0AW18 K7PQ78 A0A0F6QCS7 B4J058 B3NDC3 A0A0K8TPK2 K7PR58 A0A1A9Y261 Q2LZ16 B4GZR7 K7PR59 A0A1B0CS16 A0A3B0J420 A0A1L8E562 K7J6V1 A0A0F6QB24 E2BDQ8 A0A336LRF6 A0A0U4JS04 G0N6W0 O17953 E3N753 A0A261B5H6 A0A0J7KJN3 A0A1I7UV22 E1ZX53 A0A0U4IDU2 A0A2G5TXX6 A0A2P2HXL3 A8Y0F0 N6T3X8 E9IVN8 A0A0U4AV64 A0A195FWE8 A0A195EE33 A0A151XA98 K7PQ79 A0A1B6HDG4 A0A0F6QE70 A0A195CT09 F4WLJ5 A0A0A9XAA8 A0A026WWC0 A0A0F6QCA3 A0A1B6LXD2 A0A232EGN4 A0A0N7Z9G7 R4G4V3 A0A1S6KZB2 A0A1J1HUU2 A0A226E0Z4 A0A3P8F2V0 A0A2H2IN90 A0A1D2MWM9 A0A069DVF9 A0A310SQX7 A0A0L7RD32
A0A212FM25 A0A2H1VCT0 A0A2W1C0K5 A0A194Q332 I4DJF2 A0A2A4IX85 A0A0L0CMD7 B4L0F4 W8C1K5 T1PHL2 A0A1I8NPA9 A0A0K8TZ55 A0A034VK02 A0A0A1XNC7 B4LIF2 D3TRU9 A0A1A9URY6 A0A1L8EEI8 Q9VVL7 K7PQ56 B3M3U9 A0A1B0A6P9 A0A1L6K358 K7PR60 A0A0J9RYP2 B4HLB4 A0A1Y1L6U1 A0A1W4W542 K7PQS7 B4PJH9 A0A0M5JB65 K7PQU4 B4N4T3 A0A1L6K360 K7PQU5 K7PQ54 B4QPB7 A0A1B0AW18 K7PQ78 A0A0F6QCS7 B4J058 B3NDC3 A0A0K8TPK2 K7PR58 A0A1A9Y261 Q2LZ16 B4GZR7 K7PR59 A0A1B0CS16 A0A3B0J420 A0A1L8E562 K7J6V1 A0A0F6QB24 E2BDQ8 A0A336LRF6 A0A0U4JS04 G0N6W0 O17953 E3N753 A0A261B5H6 A0A0J7KJN3 A0A1I7UV22 E1ZX53 A0A0U4IDU2 A0A2G5TXX6 A0A2P2HXL3 A8Y0F0 N6T3X8 E9IVN8 A0A0U4AV64 A0A195FWE8 A0A195EE33 A0A151XA98 K7PQ79 A0A1B6HDG4 A0A0F6QE70 A0A195CT09 F4WLJ5 A0A0A9XAA8 A0A026WWC0 A0A0F6QCA3 A0A1B6LXD2 A0A232EGN4 A0A0N7Z9G7 R4G4V3 A0A1S6KZB2 A0A1J1HUU2 A0A226E0Z4 A0A3P8F2V0 A0A2H2IN90 A0A1D2MWM9 A0A069DVF9 A0A310SQX7 A0A0L7RD32
EC Number
1.8.1.4
Pubmed
19121390
20093189
22651552
23622113
9373151
22118469
+ More
28756777 26354079 26108605 17994087 24495485 25315136 25348373 25830018 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23139334 22936249 28004739 17550304 18057021 24059691 25853517 26369729 15632085 20075255 20798317 26774057 9851916 14624247 23537049 21282665 21719571 25401762 26823975 24508170 30249741 28648823 27129103 24330026 27289101 26334808
28756777 26354079 26108605 17994087 24495485 25315136 25348373 25830018 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23139334 22936249 28004739 17550304 18057021 24059691 25853517 26369729 15632085 20075255 20798317 26774057 9851916 14624247 23537049 21282665 21719571 25401762 26823975 24508170 30249741 28648823 27129103 24330026 27289101 26334808
EMBL
BABH01013298
BABH01013299
AF529135
AAM93255.1
HM126490
ADG45566.1
+ More
AK402493 BAM19115.1 GAIX01003839 JAA88721.1 AF008586 RSAL01000005 RVE54366.1 AGBW02007691 OWR54739.1 ODYU01001851 SOQ38627.1 KZ149892 PZC79027.1 KQ459586 KPI97815.1 AK401420 BAM18042.1 NWSH01005242 PCG64341.1 JRES01000296 KNC32634.1 CH933809 EDW19123.1 GAMC01003517 JAC03039.1 KA647378 AFP62007.1 GDHF01032746 JAI19568.1 GAKP01016148 JAC42804.1 GBXI01001766 JAD12526.1 CH940647 EDW70739.1 CCAG010008063 EZ424151 ADD20427.1 GFDG01001780 JAV17019.1 AE014296 AY058740 AAF49294.1 AAL13969.1 JX434602 AFP20528.1 CH902618 EDV39283.1 KX774473 KX774474 KX774475 KX774476 KX774477 KX774479 APR62612.1 APR62613.1 APR62614.1 APR62615.1 APR62616.1 APR62618.1 JX434596 JX434608 AFP20534.1 CM002912 KMZ00325.1 CH480815 EDW41934.1 GEZM01065180 GEZM01065179 JAV68538.1 JX434601 AFP20527.1 CM000159 EDW94667.1 CP012525 ALC43868.1 JX434600 MH891063 MH891065 MH891066 AFP20526.1 QBH99336.1 QBH99338.1 QBH99339.1 CH964095 EDW79157.1 KRF99016.1 KX774472 KX774478 APR62611.1 APR62617.1 JX434605 KF032715 AFP20531.1 AGX84981.1 JX434597 MH891069 AFP20523.1 QBH99342.1 CM000363 EDX10911.1 JXJN01004447 JX434599 AFP20525.1 KP843188 AKD42940.1 CH916366 EDV97851.1 CH954178 EDV51916.1 GDAI01001783 JAI15820.1 JX434598 AFP20524.1 CH379069 EAL29693.1 CH479199 EDW29494.1 JX434603 AFP20529.1 AJWK01025561 AJWK01025562 OUUW01000002 SPP76494.1 GFDF01000412 JAV13672.1 AAZX01011402 AAZX01011404 AAZX01017942 KP843190 AKD42942.1 GL447678 EFN86220.1 UFQT01000131 SSX20616.1 KT893322 KT893323 ALY05704.1 ALY05705.1 GL379845 EGT53974.1 Z82277 DS268545 EFO88408.1 NIPN01000162 OZG05582.1 LBMM01006632 KMQ90446.1 GL435014 EFN74243.1 KT893325 KT893326 KT893327 ALY05707.1 ALY05708.1 ALY05709.1 PDUG01000004 PIC31951.1 IACF01000746 LAB66502.1 HE601157 CAP38335.2 APGK01053269 APGK01053270 KB741223 KB632273 ENN72338.1 ERL91063.1 GL766328 EFZ15353.1 KT893324 ALY05706.1 KQ981208 KYN44751.1 KQ979039 KYN23366.1 KQ982351 KYQ57269.1 JX434604 AFP20530.1 GECU01034976 JAS72730.1 KP843191 AKD42943.1 KQ977372 KYN03264.1 GL888208 EGI64931.1 GBHO01027038 GBRD01015774 GDHC01009946 JAG16566.1 JAG50052.1 JAQ08683.1 KK107085 QOIP01000008 EZA60031.1 RLU19442.1 KP843189 AKD42941.1 GEBQ01011628 JAT28349.1 NNAY01004725 OXU17511.1 GDKW01000595 JAI56000.1 ACPB03011520 GAHY01000431 JAA77079.1 KC223159 AQT27062.1 CVRI01000021 CRK91839.1 LNIX01000008 OXA51139.1 UZAH01031137 VDP14026.1 LJIJ01000451 ODM97358.1 GBGD01001192 JAC87697.1 KQ760869 OAD58780.1 KQ414614 KOC68872.1
AK402493 BAM19115.1 GAIX01003839 JAA88721.1 AF008586 RSAL01000005 RVE54366.1 AGBW02007691 OWR54739.1 ODYU01001851 SOQ38627.1 KZ149892 PZC79027.1 KQ459586 KPI97815.1 AK401420 BAM18042.1 NWSH01005242 PCG64341.1 JRES01000296 KNC32634.1 CH933809 EDW19123.1 GAMC01003517 JAC03039.1 KA647378 AFP62007.1 GDHF01032746 JAI19568.1 GAKP01016148 JAC42804.1 GBXI01001766 JAD12526.1 CH940647 EDW70739.1 CCAG010008063 EZ424151 ADD20427.1 GFDG01001780 JAV17019.1 AE014296 AY058740 AAF49294.1 AAL13969.1 JX434602 AFP20528.1 CH902618 EDV39283.1 KX774473 KX774474 KX774475 KX774476 KX774477 KX774479 APR62612.1 APR62613.1 APR62614.1 APR62615.1 APR62616.1 APR62618.1 JX434596 JX434608 AFP20534.1 CM002912 KMZ00325.1 CH480815 EDW41934.1 GEZM01065180 GEZM01065179 JAV68538.1 JX434601 AFP20527.1 CM000159 EDW94667.1 CP012525 ALC43868.1 JX434600 MH891063 MH891065 MH891066 AFP20526.1 QBH99336.1 QBH99338.1 QBH99339.1 CH964095 EDW79157.1 KRF99016.1 KX774472 KX774478 APR62611.1 APR62617.1 JX434605 KF032715 AFP20531.1 AGX84981.1 JX434597 MH891069 AFP20523.1 QBH99342.1 CM000363 EDX10911.1 JXJN01004447 JX434599 AFP20525.1 KP843188 AKD42940.1 CH916366 EDV97851.1 CH954178 EDV51916.1 GDAI01001783 JAI15820.1 JX434598 AFP20524.1 CH379069 EAL29693.1 CH479199 EDW29494.1 JX434603 AFP20529.1 AJWK01025561 AJWK01025562 OUUW01000002 SPP76494.1 GFDF01000412 JAV13672.1 AAZX01011402 AAZX01011404 AAZX01017942 KP843190 AKD42942.1 GL447678 EFN86220.1 UFQT01000131 SSX20616.1 KT893322 KT893323 ALY05704.1 ALY05705.1 GL379845 EGT53974.1 Z82277 DS268545 EFO88408.1 NIPN01000162 OZG05582.1 LBMM01006632 KMQ90446.1 GL435014 EFN74243.1 KT893325 KT893326 KT893327 ALY05707.1 ALY05708.1 ALY05709.1 PDUG01000004 PIC31951.1 IACF01000746 LAB66502.1 HE601157 CAP38335.2 APGK01053269 APGK01053270 KB741223 KB632273 ENN72338.1 ERL91063.1 GL766328 EFZ15353.1 KT893324 ALY05706.1 KQ981208 KYN44751.1 KQ979039 KYN23366.1 KQ982351 KYQ57269.1 JX434604 AFP20530.1 GECU01034976 JAS72730.1 KP843191 AKD42943.1 KQ977372 KYN03264.1 GL888208 EGI64931.1 GBHO01027038 GBRD01015774 GDHC01009946 JAG16566.1 JAG50052.1 JAQ08683.1 KK107085 QOIP01000008 EZA60031.1 RLU19442.1 KP843189 AKD42941.1 GEBQ01011628 JAT28349.1 NNAY01004725 OXU17511.1 GDKW01000595 JAI56000.1 ACPB03011520 GAHY01000431 JAA77079.1 KC223159 AQT27062.1 CVRI01000021 CRK91839.1 LNIX01000008 OXA51139.1 UZAH01031137 VDP14026.1 LJIJ01000451 ODM97358.1 GBGD01001192 JAC87697.1 KQ760869 OAD58780.1 KQ414614 KOC68872.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000218220
UP000037069
+ More
UP000009192 UP000095301 UP000095300 UP000008792 UP000092444 UP000078200 UP000000803 UP000007801 UP000092445 UP000001292 UP000192221 UP000002282 UP000092553 UP000007798 UP000000304 UP000092460 UP000001070 UP000008711 UP000092443 UP000001819 UP000008744 UP000092461 UP000268350 UP000002358 UP000008237 UP000008068 UP000001940 UP000008281 UP000216463 UP000036403 UP000095282 UP000000311 UP000230233 UP000008549 UP000019118 UP000030742 UP000078541 UP000078492 UP000075809 UP000078542 UP000007755 UP000053097 UP000279307 UP000215335 UP000015103 UP000183832 UP000198287 UP000005237 UP000094527 UP000053825
UP000009192 UP000095301 UP000095300 UP000008792 UP000092444 UP000078200 UP000000803 UP000007801 UP000092445 UP000001292 UP000192221 UP000002282 UP000092553 UP000007798 UP000000304 UP000092460 UP000001070 UP000008711 UP000092443 UP000001819 UP000008744 UP000092461 UP000268350 UP000002358 UP000008237 UP000008068 UP000001940 UP000008281 UP000216463 UP000036403 UP000095282 UP000000311 UP000230233 UP000008549 UP000019118 UP000030742 UP000078541 UP000078492 UP000075809 UP000078542 UP000007755 UP000053097 UP000279307 UP000215335 UP000015103 UP000183832 UP000198287 UP000005237 UP000094527 UP000053825
Interpro
Gene 3D
ProteinModelPortal
Q8MUB0
D6R773
I4DMH5
S4P991
O18480
A0A3S2M9V9
+ More
A0A212FM25 A0A2H1VCT0 A0A2W1C0K5 A0A194Q332 I4DJF2 A0A2A4IX85 A0A0L0CMD7 B4L0F4 W8C1K5 T1PHL2 A0A1I8NPA9 A0A0K8TZ55 A0A034VK02 A0A0A1XNC7 B4LIF2 D3TRU9 A0A1A9URY6 A0A1L8EEI8 Q9VVL7 K7PQ56 B3M3U9 A0A1B0A6P9 A0A1L6K358 K7PR60 A0A0J9RYP2 B4HLB4 A0A1Y1L6U1 A0A1W4W542 K7PQS7 B4PJH9 A0A0M5JB65 K7PQU4 B4N4T3 A0A1L6K360 K7PQU5 K7PQ54 B4QPB7 A0A1B0AW18 K7PQ78 A0A0F6QCS7 B4J058 B3NDC3 A0A0K8TPK2 K7PR58 A0A1A9Y261 Q2LZ16 B4GZR7 K7PR59 A0A1B0CS16 A0A3B0J420 A0A1L8E562 K7J6V1 A0A0F6QB24 E2BDQ8 A0A336LRF6 A0A0U4JS04 G0N6W0 O17953 E3N753 A0A261B5H6 A0A0J7KJN3 A0A1I7UV22 E1ZX53 A0A0U4IDU2 A0A2G5TXX6 A0A2P2HXL3 A8Y0F0 N6T3X8 E9IVN8 A0A0U4AV64 A0A195FWE8 A0A195EE33 A0A151XA98 K7PQ79 A0A1B6HDG4 A0A0F6QE70 A0A195CT09 F4WLJ5 A0A0A9XAA8 A0A026WWC0 A0A0F6QCA3 A0A1B6LXD2 A0A232EGN4 A0A0N7Z9G7 R4G4V3 A0A1S6KZB2 A0A1J1HUU2 A0A226E0Z4 A0A3P8F2V0 A0A2H2IN90 A0A1D2MWM9 A0A069DVF9 A0A310SQX7 A0A0L7RD32
A0A212FM25 A0A2H1VCT0 A0A2W1C0K5 A0A194Q332 I4DJF2 A0A2A4IX85 A0A0L0CMD7 B4L0F4 W8C1K5 T1PHL2 A0A1I8NPA9 A0A0K8TZ55 A0A034VK02 A0A0A1XNC7 B4LIF2 D3TRU9 A0A1A9URY6 A0A1L8EEI8 Q9VVL7 K7PQ56 B3M3U9 A0A1B0A6P9 A0A1L6K358 K7PR60 A0A0J9RYP2 B4HLB4 A0A1Y1L6U1 A0A1W4W542 K7PQS7 B4PJH9 A0A0M5JB65 K7PQU4 B4N4T3 A0A1L6K360 K7PQU5 K7PQ54 B4QPB7 A0A1B0AW18 K7PQ78 A0A0F6QCS7 B4J058 B3NDC3 A0A0K8TPK2 K7PR58 A0A1A9Y261 Q2LZ16 B4GZR7 K7PR59 A0A1B0CS16 A0A3B0J420 A0A1L8E562 K7J6V1 A0A0F6QB24 E2BDQ8 A0A336LRF6 A0A0U4JS04 G0N6W0 O17953 E3N753 A0A261B5H6 A0A0J7KJN3 A0A1I7UV22 E1ZX53 A0A0U4IDU2 A0A2G5TXX6 A0A2P2HXL3 A8Y0F0 N6T3X8 E9IVN8 A0A0U4AV64 A0A195FWE8 A0A195EE33 A0A151XA98 K7PQ79 A0A1B6HDG4 A0A0F6QE70 A0A195CT09 F4WLJ5 A0A0A9XAA8 A0A026WWC0 A0A0F6QCA3 A0A1B6LXD2 A0A232EGN4 A0A0N7Z9G7 R4G4V3 A0A1S6KZB2 A0A1J1HUU2 A0A226E0Z4 A0A3P8F2V0 A0A2H2IN90 A0A1D2MWM9 A0A069DVF9 A0A310SQX7 A0A0L7RD32
PDB
5NHG
E-value=1.67923e-176,
Score=1591
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
00020 Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00260 Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
00020 Citrate cycle (TCA cycle) - Bombyx mori (domestic silkworm)
00260 Glycine, serine and threonine metabolism - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
GO
Topology
Subcellular location
Cytoplasm
Mitochondrion matrix
Mitochondrion matrix
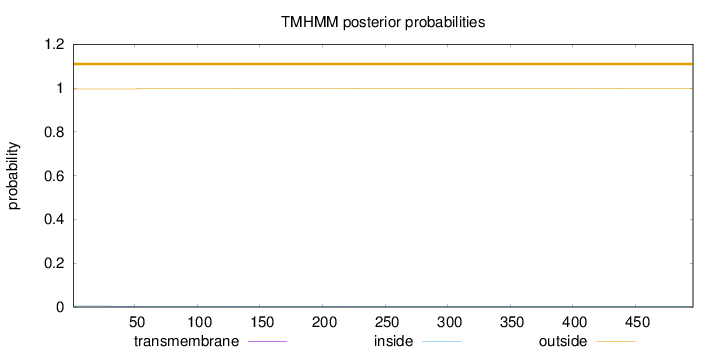
Length:
496
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0750400000000001
Exp number, first 60 AAs:
0.06187
Total prob of N-in:
0.00449
outside
1 - 496
Population Genetic Test Statistics
Pi
27.997291
Theta
21.918366
Tajima's D
1.096278
CLR
1.5704
CSRT
0.694065296735163
Interpretation
Uncertain
