Gene
KWMTBOMO13036
Pre Gene Modal
BGIBMGA000420
Annotation
PREDICTED:_serine/threonine_kinase_LKB1_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.781 Nuclear Reliability : 2.479
Sequence
CDS
ATGGTTGGAAAGTATGTCATGGGTGATGTTTTAGGAGAAGGTTCATACGGAAAAGTCAAAGAAATGTTAGACTCGCAATCTTTATGTAGGCGAGCCGTAAAAATTCTTAAAAAGAGGAAATTAAGAAGAATACCAAATGGTGAACAAAATGTACAGAGAGAGATACAATTACTAAGAATCCTTAGACATAAAAATGTGATAGAACTAGTCGATGTTATCTATAATGATGAGAAACAAAAGATGTATCTTGTTATGGAATTCTGTGTTGGAGTATTACAGGACATGCTGGAGTCAAGTCCCGGTAAAAAGTTTCCTCAAAGACAAGCTCATGATTACTTTCTACAGCTGCTGGACGGATTAGATTACTTACATGGGCAGGGTGTTGTACATAAAGATATTAAACCAGGAAATTTACTTCTGACACTGGATCAAACTCTCAAAATCACAGATTTTGGTGTTGCTGAGGCATTAGACATGTTTTCACAGGAGGATATATGTTACACAGGACAGGGCTCACCTGCATTCCAGCCTCCAGAGATAGCCAATGGTGCTGAACAATTTTCTGGAACTAAAGTTGATATATGGAGCAGTGGAGTTACATTGTACAACATGACAACAGGGAGGTATCCGTTCGAGGGTGACAACGTGTACCGATTGCTGGAGGCGATAAGCCGGGGTCCCCCCGCGCCCCCGCCCGCGAGCGTGGGCCCCGCTCTCAGCGCTCTGCTCGTGCACATGCTCAACCGAGACCCTACACTAAGACCCACCGTCCACGAGATCAGGAGACACGCATGGGTGCAGGCAACACCGCCCTTCGAACCGGGCGAGAAACTAGTGAATCCGAGATCTCGACGTTCCGACGAGCTTCACACTTCGACTGTAATACCGTATCTCGTCGAGAGGCATTACGCTTCTGCGCAGGAATACTTCACCGAAAGGGACCTGCGGCAAGGGCTGGGTGACGGCGGCTCCCGGACGATGTCGACTGCGGAATTGTCTGACGGAGGATCGGGGCAGCACAAGAGGCGATGGCACTCGTCGTGCGGCCGGCTGCCCTCGTGCCGGCTCACGTGA
Protein
MVGKYVMGDVLGEGSYGKVKEMLDSQSLCRRAVKILKKRKLRRIPNGEQNVQREIQLLRILRHKNVIELVDVIYNDEKQKMYLVMEFCVGVLQDMLESSPGKKFPQRQAHDYFLQLLDGLDYLHGQGVVHKDIKPGNLLLTLDQTLKITDFGVAEALDMFSQEDICYTGQGSPAFQPPEIANGAEQFSGTKVDIWSSGVTLYNMTTGRYPFEGDNVYRLLEAISRGPPAPPPASVGPALSALLVHMLNRDPTLRPTVHEIRRHAWVQATPPFEPGEKLVNPRSRRSDELHTSTVIPYLVERHYASAQEYFTERDLRQGLGDGGSRTMSTAELSDGGSGQHKRRWHSSCGRLPSCRLT
Summary
Similarity
Belongs to the protein kinase superfamily.
Uniprot
A0A3S2NM10
A0A2A4J7X3
A0A2W1BVA9
A0A2H1VHV5
A0A194QW25
H9IT41
+ More
A0A194PWS1 A0A212F481 A0A2J7QVW7 A0A158P322 E2AJH8 F4WM76 A0A067R0B3 A0A2P8Z3J3 A0A0A9XWP5 A0A1B0CEH8 A0A1B0DPW2 A0A2R7W989 A0A1W4WL10 A0A195BBZ2 A0A1W4WAY1 A0A026WAI1 A0A195DCC3 A0A151IGF1 A0A1Y1K0R7 A0A0P4VJT5 R4G8A9 T1HN05 A0A1B6L8B9 A0A1L8DKA3 K7J666 A0A088APS1 A0A232F6V5 A0A023F908 A0A069DTU5 A0A1B6EF04 A0A0V0GAQ1 A0A224XKD1 A0A0L7QQA4 A0A0L7L7A0 A0A0M9A2H1 E9IEY0 A0A2A3E500 A0A154PFV9 A0A0L0CEI5 A0A1I8NZ80 D6X0P9 A0A195F3K0 A0A0M4MF00 A0A1I8M4E4 T1PIL4 A0A1A9W605 A0A182JB42 A0A1B6HBW0 A0A182GXB7 D3TMT6 A0A1A9XJ21 A0A1B0BK01 A0A182QVN6 A0A182N4L3 A0A293M950 A0A293MY28 A0A182P579 A0A2J7QVW5 A0A182JQC8 A0A084WTF2 A0A1A9V749 A0A1A9Z488 A0A1B0FGP7 A0A2M4BIN5 A0A2M4BIM8 A0A182XE76 A0A182TTQ6 Q7QBI7 A0A182HJ47 A0A182RPM6 W5JFF4 A0A182FR88 A0A2S2QL85 A0A2M4CZ85 E0W1E5 B3LXA9 W8CAC6 B3P420 A0A182Y7B4 A0A0P8XX91 N6U5C4 B4PQH4 A0A1Q3FAE3 A0A1E1XF88 A0A1Q3FB09 B4M0X1 A0A1Q3F9U9 Q297P6 A0A1Q3FAZ8 A0A1W4V4U5 A0A2H8TMY0 Q178C1 B4G2U3
A0A194PWS1 A0A212F481 A0A2J7QVW7 A0A158P322 E2AJH8 F4WM76 A0A067R0B3 A0A2P8Z3J3 A0A0A9XWP5 A0A1B0CEH8 A0A1B0DPW2 A0A2R7W989 A0A1W4WL10 A0A195BBZ2 A0A1W4WAY1 A0A026WAI1 A0A195DCC3 A0A151IGF1 A0A1Y1K0R7 A0A0P4VJT5 R4G8A9 T1HN05 A0A1B6L8B9 A0A1L8DKA3 K7J666 A0A088APS1 A0A232F6V5 A0A023F908 A0A069DTU5 A0A1B6EF04 A0A0V0GAQ1 A0A224XKD1 A0A0L7QQA4 A0A0L7L7A0 A0A0M9A2H1 E9IEY0 A0A2A3E500 A0A154PFV9 A0A0L0CEI5 A0A1I8NZ80 D6X0P9 A0A195F3K0 A0A0M4MF00 A0A1I8M4E4 T1PIL4 A0A1A9W605 A0A182JB42 A0A1B6HBW0 A0A182GXB7 D3TMT6 A0A1A9XJ21 A0A1B0BK01 A0A182QVN6 A0A182N4L3 A0A293M950 A0A293MY28 A0A182P579 A0A2J7QVW5 A0A182JQC8 A0A084WTF2 A0A1A9V749 A0A1A9Z488 A0A1B0FGP7 A0A2M4BIN5 A0A2M4BIM8 A0A182XE76 A0A182TTQ6 Q7QBI7 A0A182HJ47 A0A182RPM6 W5JFF4 A0A182FR88 A0A2S2QL85 A0A2M4CZ85 E0W1E5 B3LXA9 W8CAC6 B3P420 A0A182Y7B4 A0A0P8XX91 N6U5C4 B4PQH4 A0A1Q3FAE3 A0A1E1XF88 A0A1Q3FB09 B4M0X1 A0A1Q3F9U9 Q297P6 A0A1Q3FAZ8 A0A1W4V4U5 A0A2H8TMY0 Q178C1 B4G2U3
Pubmed
28756777
26354079
19121390
22118469
21347285
20798317
+ More
21719571 24845553 29403074 25401762 26823975 24508170 30249741 28004739 27129103 20075255 28648823 25474469 26334808 26227816 21282665 26108605 18362917 19820115 25315136 26483478 20353571 24438588 12364791 14747013 17210077 20920257 23761445 20566863 17994087 24495485 25244985 23537049 17550304 28503490 15632085 17510324
21719571 24845553 29403074 25401762 26823975 24508170 30249741 28004739 27129103 20075255 28648823 25474469 26334808 26227816 21282665 26108605 18362917 19820115 25315136 26483478 20353571 24438588 12364791 14747013 17210077 20920257 23761445 20566863 17994087 24495485 25244985 23537049 17550304 28503490 15632085 17510324
EMBL
RSAL01000005
RVE54371.1
NWSH01002803
PCG67502.1
KZ149892
PZC79022.1
+ More
ODYU01002641 SOQ40409.1 KQ461073 KPJ09738.1 BABH01013306 KQ459586 KPI97811.1 AGBW02010421 OWR48540.1 NEVH01009770 PNF32712.1 ADTU01001302 GL440027 EFN66373.1 GL888217 EGI64720.1 KK852801 KDR16301.1 PYGN01000212 PSN51071.1 GBHO01018387 GBRD01003536 GDHC01004496 JAG25217.1 JAG62285.1 JAQ14133.1 AJWK01008878 AJWK01008879 AJVK01018648 AJVK01018649 KK854482 PTY16253.1 KQ976522 KYM82081.1 KK107356 QOIP01000010 EZA52039.1 RLU17468.1 KQ980989 KYN10560.1 KQ977726 KYN00288.1 GEZM01101726 JAV52357.1 GDKW01002096 JAI54499.1 GAHY01001292 JAA76218.1 ACPB03015113 GEBQ01020014 JAT19963.1 GFDF01007191 JAV06893.1 NNAY01000881 OXU26087.1 GBBI01000765 JAC17947.1 GBGD01001549 JAC87340.1 GEDC01000808 JAS36490.1 GECL01001268 JAP04856.1 GFTR01006148 JAW10278.1 KQ414786 KOC60818.1 JTDY01002539 KOB71219.1 KQ435789 KOX74479.1 GL762728 EFZ20869.1 KZ288391 PBC26362.1 KQ434896 KZC10735.1 JRES01000501 KNC30670.1 KQ971371 EFA10720.1 KQ981855 KYN34757.1 KR075853 ALE20572.1 KA648611 AFP63240.1 GECU01035536 GECU01012394 JAS72170.1 JAS95312.1 JXUM01095090 JXUM01095091 JXUM01095092 KQ564172 KXJ72635.1 EZ422738 ADD19014.1 JXJN01015758 AXCN02000045 GFWV01012585 MAA37314.1 GFWV01021024 MAA45752.1 PNF32714.1 ATLV01026890 KE525420 KFB53496.1 CCAG010002945 GGFJ01003774 MBW52915.1 GGFJ01003775 MBW52916.1 AAAB01008879 EAA08443.5 APCN01002124 ADMH02001656 ETN61539.1 GGMS01009313 MBY78516.1 GGFL01006466 MBW70644.1 AAZO01007007 DS235870 EEB19451.1 CH902617 EDV41709.1 GAMC01005876 JAC00680.1 CH954181 EDV49194.1 KPU79306.1 APGK01041758 KB740997 KB632100 ENN75846.1 ERL88845.1 CM000160 EDW97270.1 KRK03614.1 KRK03615.1 GFDL01010567 JAV24478.1 GFAC01001283 JAT97905.1 GFDL01010357 JAV24688.1 CH940650 EDW67382.2 GFDL01010691 JAV24354.1 CM000070 EAL28159.2 GFDL01010346 JAV24699.1 GFXV01003680 MBW15485.1 CH477366 EAT42532.1 CH479179 EDW24138.1
ODYU01002641 SOQ40409.1 KQ461073 KPJ09738.1 BABH01013306 KQ459586 KPI97811.1 AGBW02010421 OWR48540.1 NEVH01009770 PNF32712.1 ADTU01001302 GL440027 EFN66373.1 GL888217 EGI64720.1 KK852801 KDR16301.1 PYGN01000212 PSN51071.1 GBHO01018387 GBRD01003536 GDHC01004496 JAG25217.1 JAG62285.1 JAQ14133.1 AJWK01008878 AJWK01008879 AJVK01018648 AJVK01018649 KK854482 PTY16253.1 KQ976522 KYM82081.1 KK107356 QOIP01000010 EZA52039.1 RLU17468.1 KQ980989 KYN10560.1 KQ977726 KYN00288.1 GEZM01101726 JAV52357.1 GDKW01002096 JAI54499.1 GAHY01001292 JAA76218.1 ACPB03015113 GEBQ01020014 JAT19963.1 GFDF01007191 JAV06893.1 NNAY01000881 OXU26087.1 GBBI01000765 JAC17947.1 GBGD01001549 JAC87340.1 GEDC01000808 JAS36490.1 GECL01001268 JAP04856.1 GFTR01006148 JAW10278.1 KQ414786 KOC60818.1 JTDY01002539 KOB71219.1 KQ435789 KOX74479.1 GL762728 EFZ20869.1 KZ288391 PBC26362.1 KQ434896 KZC10735.1 JRES01000501 KNC30670.1 KQ971371 EFA10720.1 KQ981855 KYN34757.1 KR075853 ALE20572.1 KA648611 AFP63240.1 GECU01035536 GECU01012394 JAS72170.1 JAS95312.1 JXUM01095090 JXUM01095091 JXUM01095092 KQ564172 KXJ72635.1 EZ422738 ADD19014.1 JXJN01015758 AXCN02000045 GFWV01012585 MAA37314.1 GFWV01021024 MAA45752.1 PNF32714.1 ATLV01026890 KE525420 KFB53496.1 CCAG010002945 GGFJ01003774 MBW52915.1 GGFJ01003775 MBW52916.1 AAAB01008879 EAA08443.5 APCN01002124 ADMH02001656 ETN61539.1 GGMS01009313 MBY78516.1 GGFL01006466 MBW70644.1 AAZO01007007 DS235870 EEB19451.1 CH902617 EDV41709.1 GAMC01005876 JAC00680.1 CH954181 EDV49194.1 KPU79306.1 APGK01041758 KB740997 KB632100 ENN75846.1 ERL88845.1 CM000160 EDW97270.1 KRK03614.1 KRK03615.1 GFDL01010567 JAV24478.1 GFAC01001283 JAT97905.1 GFDL01010357 JAV24688.1 CH940650 EDW67382.2 GFDL01010691 JAV24354.1 CM000070 EAL28159.2 GFDL01010346 JAV24699.1 GFXV01003680 MBW15485.1 CH477366 EAT42532.1 CH479179 EDW24138.1
Proteomes
UP000283053
UP000218220
UP000053240
UP000005204
UP000053268
UP000007151
+ More
UP000235965 UP000005205 UP000000311 UP000007755 UP000027135 UP000245037 UP000092461 UP000092462 UP000192223 UP000078540 UP000053097 UP000279307 UP000078492 UP000078542 UP000015103 UP000002358 UP000005203 UP000215335 UP000053825 UP000037510 UP000053105 UP000242457 UP000076502 UP000037069 UP000095300 UP000007266 UP000078541 UP000095301 UP000091820 UP000075880 UP000069940 UP000249989 UP000092443 UP000092460 UP000075886 UP000075884 UP000075885 UP000075881 UP000030765 UP000078200 UP000092445 UP000092444 UP000076407 UP000075902 UP000007062 UP000075840 UP000075900 UP000000673 UP000069272 UP000009046 UP000007801 UP000008711 UP000076408 UP000019118 UP000030742 UP000002282 UP000008792 UP000001819 UP000192221 UP000008820 UP000008744
UP000235965 UP000005205 UP000000311 UP000007755 UP000027135 UP000245037 UP000092461 UP000092462 UP000192223 UP000078540 UP000053097 UP000279307 UP000078492 UP000078542 UP000015103 UP000002358 UP000005203 UP000215335 UP000053825 UP000037510 UP000053105 UP000242457 UP000076502 UP000037069 UP000095300 UP000007266 UP000078541 UP000095301 UP000091820 UP000075880 UP000069940 UP000249989 UP000092443 UP000092460 UP000075886 UP000075884 UP000075885 UP000075881 UP000030765 UP000078200 UP000092445 UP000092444 UP000076407 UP000075902 UP000007062 UP000075840 UP000075900 UP000000673 UP000069272 UP000009046 UP000007801 UP000008711 UP000076408 UP000019118 UP000030742 UP000002282 UP000008792 UP000001819 UP000192221 UP000008820 UP000008744
PRIDE
Pfam
PF00069 Pkinase
Interpro
SUPFAM
SSF56112
SSF56112
ProteinModelPortal
A0A3S2NM10
A0A2A4J7X3
A0A2W1BVA9
A0A2H1VHV5
A0A194QW25
H9IT41
+ More
A0A194PWS1 A0A212F481 A0A2J7QVW7 A0A158P322 E2AJH8 F4WM76 A0A067R0B3 A0A2P8Z3J3 A0A0A9XWP5 A0A1B0CEH8 A0A1B0DPW2 A0A2R7W989 A0A1W4WL10 A0A195BBZ2 A0A1W4WAY1 A0A026WAI1 A0A195DCC3 A0A151IGF1 A0A1Y1K0R7 A0A0P4VJT5 R4G8A9 T1HN05 A0A1B6L8B9 A0A1L8DKA3 K7J666 A0A088APS1 A0A232F6V5 A0A023F908 A0A069DTU5 A0A1B6EF04 A0A0V0GAQ1 A0A224XKD1 A0A0L7QQA4 A0A0L7L7A0 A0A0M9A2H1 E9IEY0 A0A2A3E500 A0A154PFV9 A0A0L0CEI5 A0A1I8NZ80 D6X0P9 A0A195F3K0 A0A0M4MF00 A0A1I8M4E4 T1PIL4 A0A1A9W605 A0A182JB42 A0A1B6HBW0 A0A182GXB7 D3TMT6 A0A1A9XJ21 A0A1B0BK01 A0A182QVN6 A0A182N4L3 A0A293M950 A0A293MY28 A0A182P579 A0A2J7QVW5 A0A182JQC8 A0A084WTF2 A0A1A9V749 A0A1A9Z488 A0A1B0FGP7 A0A2M4BIN5 A0A2M4BIM8 A0A182XE76 A0A182TTQ6 Q7QBI7 A0A182HJ47 A0A182RPM6 W5JFF4 A0A182FR88 A0A2S2QL85 A0A2M4CZ85 E0W1E5 B3LXA9 W8CAC6 B3P420 A0A182Y7B4 A0A0P8XX91 N6U5C4 B4PQH4 A0A1Q3FAE3 A0A1E1XF88 A0A1Q3FB09 B4M0X1 A0A1Q3F9U9 Q297P6 A0A1Q3FAZ8 A0A1W4V4U5 A0A2H8TMY0 Q178C1 B4G2U3
A0A194PWS1 A0A212F481 A0A2J7QVW7 A0A158P322 E2AJH8 F4WM76 A0A067R0B3 A0A2P8Z3J3 A0A0A9XWP5 A0A1B0CEH8 A0A1B0DPW2 A0A2R7W989 A0A1W4WL10 A0A195BBZ2 A0A1W4WAY1 A0A026WAI1 A0A195DCC3 A0A151IGF1 A0A1Y1K0R7 A0A0P4VJT5 R4G8A9 T1HN05 A0A1B6L8B9 A0A1L8DKA3 K7J666 A0A088APS1 A0A232F6V5 A0A023F908 A0A069DTU5 A0A1B6EF04 A0A0V0GAQ1 A0A224XKD1 A0A0L7QQA4 A0A0L7L7A0 A0A0M9A2H1 E9IEY0 A0A2A3E500 A0A154PFV9 A0A0L0CEI5 A0A1I8NZ80 D6X0P9 A0A195F3K0 A0A0M4MF00 A0A1I8M4E4 T1PIL4 A0A1A9W605 A0A182JB42 A0A1B6HBW0 A0A182GXB7 D3TMT6 A0A1A9XJ21 A0A1B0BK01 A0A182QVN6 A0A182N4L3 A0A293M950 A0A293MY28 A0A182P579 A0A2J7QVW5 A0A182JQC8 A0A084WTF2 A0A1A9V749 A0A1A9Z488 A0A1B0FGP7 A0A2M4BIN5 A0A2M4BIM8 A0A182XE76 A0A182TTQ6 Q7QBI7 A0A182HJ47 A0A182RPM6 W5JFF4 A0A182FR88 A0A2S2QL85 A0A2M4CZ85 E0W1E5 B3LXA9 W8CAC6 B3P420 A0A182Y7B4 A0A0P8XX91 N6U5C4 B4PQH4 A0A1Q3FAE3 A0A1E1XF88 A0A1Q3FB09 B4M0X1 A0A1Q3F9U9 Q297P6 A0A1Q3FAZ8 A0A1W4V4U5 A0A2H8TMY0 Q178C1 B4G2U3
PDB
2WTK
E-value=1.00048e-96,
Score=902
Ontologies
PATHWAY
GO
GO:0004674
GO:0005524
GO:0030295
GO:0001558
GO:0030010
GO:0042593
GO:0004672
GO:0016021
GO:0006468
GO:0005737
GO:0005634
GO:0035556
GO:0042770
GO:0044773
GO:0072697
GO:0045451
GO:0005768
GO:0005938
GO:0046328
GO:0007052
GO:0018107
GO:0045201
GO:0016325
GO:0030709
GO:0043065
GO:0010897
GO:0007391
GO:0008340
GO:0043565
GO:0007155
GO:0005515
GO:0008237
GO:0008152
GO:0016491
GO:0015930
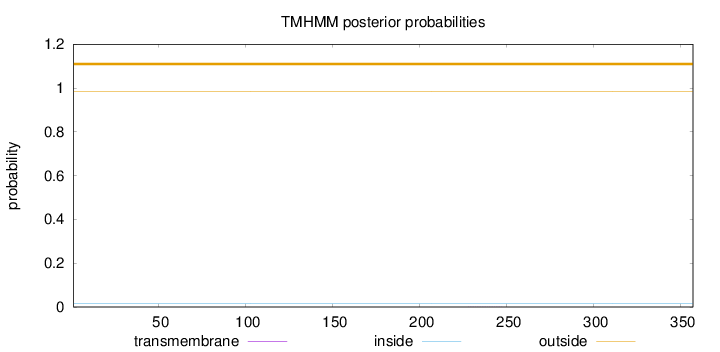
Topology
Length:
357
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00625
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01461
outside
1 - 357
Population Genetic Test Statistics
Pi
241.983344
Theta
162.615404
Tajima's D
1.223175
CLR
0.461557
CSRT
0.71896405179741
Interpretation
Uncertain
