Gene
KWMTBOMO13034
Pre Gene Modal
BGIBMGA000291
Annotation
PREDICTED:_elongation_of_very_long_chain_fatty_acids_protein_AAEL008004-like_[Amyelois_transitella]
Full name
Elongation of very long chain fatty acids protein
Alternative Name
Very-long-chain 3-oxoacyl-CoA synthase
Location in the cell
PlasmaMembrane Reliability : 4.798
Sequence
CDS
ATGGAAGCGCTGAGTAGGCTTGTGGCGGGCTACCATGACCTGATGGATAACAAGAGCGATCAAAGAGTCAAAGATTGGTTCCTCATGTCCTCACCCTTCCCCACCTTGGCGATATGTCTTACATACGTCTTCATCGTCAAAGTACTTGGACCGAAGCTAATGGAAAATAGGAAACCATTCGAATTGAAGCAAGTTCTCATTTGGTATAACTTGTTCCAAGTTATATTCAGCTGTTGGCTCTTTTATGAGAGTATAGTCAGTGGGTGGTTCACGACGTATAGTTTCCGATGTCAACCAGTGGACTATTCAAACACTCCAGAAACATTACGGACCGTCAACGGCTGTTGGTGGTATTACTTCTCGAAGTTTACAGAGTTCTTTGACACGTTCTTCTTCGTGATGCGCAAGAAGTTTGATCATGTGTCAAAGCTGCACGTCATTCACCACGGTATCATGCCAATGTCCGTTTGGTTTGGCGTCAAATTTACACCTGGTGGTCACTCCACATTCTTCGGTATGCTCAATACTTTCGTACACATCGTAATGTATTCATATTACTTGCTGGCAGCCCTCGGACCGCAGGTTCAGAAGTATCTGTGGTGGAAGAAATACCTAACCGCTCTTCAAATGGTCCAGTTCGTACTCGTGTTCATTCATGCGTTCCAGCTACTGTTCATCGACTGCGATTATCCACGAGCATTCGTTTGGTGGATCGGCATGCACGCCGTACTCTTCTACTATCTGTTTTCTGATTTCTACAAACAGGCCTACCTCAGGAAGAGCAAGAAGGCCAAATCTATGATGAAAGTGGAGGACGAACCAGTCAAGTATGAGACAAAGGAGATGAAGCTTCCACTGCTGAACGGTTTCAGCAATGGCTCAGCACTTGGTGACGTTCGGCAAAGGATGGCGGGCGCCGTGGCTTCACAGTGA
Protein
MEALSRLVAGYHDLMDNKSDQRVKDWFLMSSPFPTLAICLTYVFIVKVLGPKLMENRKPFELKQVLIWYNLFQVIFSCWLFYESIVSGWFTTYSFRCQPVDYSNTPETLRTVNGCWWYYFSKFTEFFDTFFFVMRKKFDHVSKLHVIHHGIMPMSVWFGVKFTPGGHSTFFGMLNTFVHIVMYSYYLLAALGPQVQKYLWWKKYLTALQMVQFVLVFIHAFQLLFIDCDYPRAFVWWIGMHAVLFYYLFSDFYKQAYLRKSKKAKSMMKVEDEPVKYETKEMKLPLLNGFSNGSALGDVRQRMAGAVASQ
Summary
Catalytic Activity
a very-long-chain acyl-CoA + H(+) + malonyl-CoA = a very-long-chain 3-oxoacyl-CoA + CO2 + CoA
Similarity
Belongs to the ELO family.
Uniprot
A0A2H1V9U8
A0A3S2LQ19
A0A194QXQ3
A0A194PYI3
A0A2W1C1M5
A0A212EXC4
+ More
H9ISR4 I4DP69 A0A1E1W2F2 A0A1B6FP30 A0A1B6I0V4 A0A069DYP9 A0A290Y1Y8 A0A1B6EKQ3 A0A0T6BBD3 K7J7Y5 A0A232FGG0 A0A224XJ95 D6X366 A0A1W4WQD4 A0A0V0GA67 A0A0J7KVR6 J3JXW1 A0A2J7QVS3 A0A195EDU0 A0A195FWF1 A0A3S2TCH4 A0A151XA48 A0A1B6CS52 A0A067R7D7 A0A310S8Q7 A0A1B6KPQ9 A0A1Y1L5C0 A0A3Q8FM39 F4WLM2 A0A0L7RDI8 E2C8Q7 A0A0M9A788 A0A2H1V4D9 A0A158NN79 A0A3Q8G639 E0W1G7 A0A195AV15 A0A0C9RDQ9 T1HZW8 A0A195CR72 A0A2W1BVD6 A0A146LEX3 A0A158NS34 K7J7Y3 A0A1I8M815 A0A1E1W7T1 A0A194QW86 A0A1Y1M7G3 A0A232F9G3 A0A1B6EK88 A0A194PWR4 A0A1I8PE65 A0A0R3NHN9 A0A2A3EM15 V9IJ65 A0A1L8EHX7 A0A0C9RVW1 B4NKN1 A0A0P8YFQ5 W8BLU2 A0A034VF14 A0A1I8P5I0 A0A182YJR1 A0A1I8PDW3 A0A1S3D285 B4G2U1 A0A026WVC6 A0A1B6FBI1 Q297P4 A0A1I8MHJ2 A0A1W4VLW0 A0A182RI52 A0A0K8URN1 A0A0A1XHP0 A0A1Y1M310 A0A2R7VWM8 B3M241 A0A0R3NLI7 A0A182NCS1 A0A0V0GAQ6 A0A034VK19 A0A212EXH7 A0A034VHB6 A0A0P8YNC0 A0A1L8DYF1 W8B0N6 A0A2S2QZZ1 A0A3B0K0S6 A0A1I8P5B7 A0A034VTC3 A0A1W4VZ58 A0A0K8VUG2 A0A0A9WPG5 A0A034VQQ5
H9ISR4 I4DP69 A0A1E1W2F2 A0A1B6FP30 A0A1B6I0V4 A0A069DYP9 A0A290Y1Y8 A0A1B6EKQ3 A0A0T6BBD3 K7J7Y5 A0A232FGG0 A0A224XJ95 D6X366 A0A1W4WQD4 A0A0V0GA67 A0A0J7KVR6 J3JXW1 A0A2J7QVS3 A0A195EDU0 A0A195FWF1 A0A3S2TCH4 A0A151XA48 A0A1B6CS52 A0A067R7D7 A0A310S8Q7 A0A1B6KPQ9 A0A1Y1L5C0 A0A3Q8FM39 F4WLM2 A0A0L7RDI8 E2C8Q7 A0A0M9A788 A0A2H1V4D9 A0A158NN79 A0A3Q8G639 E0W1G7 A0A195AV15 A0A0C9RDQ9 T1HZW8 A0A195CR72 A0A2W1BVD6 A0A146LEX3 A0A158NS34 K7J7Y3 A0A1I8M815 A0A1E1W7T1 A0A194QW86 A0A1Y1M7G3 A0A232F9G3 A0A1B6EK88 A0A194PWR4 A0A1I8PE65 A0A0R3NHN9 A0A2A3EM15 V9IJ65 A0A1L8EHX7 A0A0C9RVW1 B4NKN1 A0A0P8YFQ5 W8BLU2 A0A034VF14 A0A1I8P5I0 A0A182YJR1 A0A1I8PDW3 A0A1S3D285 B4G2U1 A0A026WVC6 A0A1B6FBI1 Q297P4 A0A1I8MHJ2 A0A1W4VLW0 A0A182RI52 A0A0K8URN1 A0A0A1XHP0 A0A1Y1M310 A0A2R7VWM8 B3M241 A0A0R3NLI7 A0A182NCS1 A0A0V0GAQ6 A0A034VK19 A0A212EXH7 A0A034VHB6 A0A0P8YNC0 A0A1L8DYF1 W8B0N6 A0A2S2QZZ1 A0A3B0K0S6 A0A1I8P5B7 A0A034VTC3 A0A1W4VZ58 A0A0K8VUG2 A0A0A9WPG5 A0A034VQQ5
EC Number
2.3.1.199
Pubmed
EMBL
ODYU01001237
SOQ37172.1
RSAL01000386
RVE42030.1
KQ461073
KPJ09740.1
+ More
KQ459586 KPI97809.1 KZ149892 PZC79020.1 AGBW02011787 OWR46146.1 BABH01013309 BABH01013310 BABH01013311 AK403449 BAM19709.1 GDQN01009897 GDQN01000875 JAT81157.1 JAT90179.1 GECZ01017811 JAS51958.1 GECU01027147 JAS80559.1 GBGD01002010 JAC86879.1 MF279189 ATD85029.1 GECZ01031249 JAS38520.1 LJIG01002323 KRT84617.1 NNAY01000231 OXU29834.1 GFTR01005302 JAW11124.1 KQ971372 EFA09795.1 GECL01001821 JAP04303.1 LBMM01002758 KMQ94406.1 BT128083 AEE63044.1 NEVH01009772 PNF32683.1 KQ979039 KYN23390.1 KQ981208 KYN44773.1 RVE42029.1 KQ982351 KYQ57245.1 GEDC01021034 JAS16264.1 KK852652 KDR19399.1 KQ768821 OAD53048.1 GEBQ01026570 JAT13407.1 GEZM01064008 JAV68873.1 MG573180 AWJ25042.1 GL888208 EGI64958.1 KQ414614 KOC68913.1 GL453671 EFN75681.1 KQ435720 KOX78657.1 ODYU01000613 SOQ35703.1 ADTU01021133 MG573184 AWJ25046.1 DS235870 EEB19473.1 KQ976738 KYM75897.1 GBYB01006465 JAG76232.1 ACPB03000870 KQ977372 KYN03238.1 PZC79018.1 GDHC01011928 JAQ06701.1 ADTU01024445 ADTU01024446 ADTU01024447 ADTU01024448 ADTU01024449 ADTU01024450 ADTU01024451 ADTU01024452 GDQN01007992 GDQN01000519 JAT83062.1 JAT90535.1 KPJ09742.1 GEZM01041641 JAV80245.1 NNAY01000641 OXU27242.1 GECZ01031419 GECZ01031175 GECZ01028076 JAS38350.1 JAS38594.1 JAS41693.1 KPI97806.1 CM000070 KRT00510.1 KZ288222 PBC32081.1 JR047921 AEY60541.1 GFDG01000482 JAV18317.1 GBYB01013095 JAG82862.1 CH964272 EDW84092.1 CH902617 KPU80229.1 GAMC01015951 JAB90604.1 GAKP01017051 JAC41901.1 CH479179 EDW24136.1 KK107085 QOIP01000008 EZA60005.1 RLU19769.1 GECZ01022288 JAS47481.1 EAL28161.4 GDHF01023098 JAI29216.1 GBXI01017108 GBXI01014026 GBXI01003791 GBXI01000224 JAC97183.1 JAD00266.1 JAD10501.1 JAD14068.1 GEZM01041642 JAV80244.1 KK854125 PTY11739.1 EDV43365.1 KRT00511.1 GECL01001006 JAP05118.1 GAKP01017057 GAKP01017054 GAKP01017052 JAC41898.1 OWR46144.1 GAKP01017053 JAC41899.1 KPU80280.1 GFDF01002638 JAV11446.1 GAMC01015954 GAMC01015952 JAB90603.1 GGMS01014134 MBY83337.1 OUUW01000013 SPP87834.1 GAKP01014169 JAC44783.1 GDHF01010129 JAI42185.1 GBHO01033985 GDHC01002901 JAG09619.1 JAQ15728.1 GAKP01014168 JAC44784.1
KQ459586 KPI97809.1 KZ149892 PZC79020.1 AGBW02011787 OWR46146.1 BABH01013309 BABH01013310 BABH01013311 AK403449 BAM19709.1 GDQN01009897 GDQN01000875 JAT81157.1 JAT90179.1 GECZ01017811 JAS51958.1 GECU01027147 JAS80559.1 GBGD01002010 JAC86879.1 MF279189 ATD85029.1 GECZ01031249 JAS38520.1 LJIG01002323 KRT84617.1 NNAY01000231 OXU29834.1 GFTR01005302 JAW11124.1 KQ971372 EFA09795.1 GECL01001821 JAP04303.1 LBMM01002758 KMQ94406.1 BT128083 AEE63044.1 NEVH01009772 PNF32683.1 KQ979039 KYN23390.1 KQ981208 KYN44773.1 RVE42029.1 KQ982351 KYQ57245.1 GEDC01021034 JAS16264.1 KK852652 KDR19399.1 KQ768821 OAD53048.1 GEBQ01026570 JAT13407.1 GEZM01064008 JAV68873.1 MG573180 AWJ25042.1 GL888208 EGI64958.1 KQ414614 KOC68913.1 GL453671 EFN75681.1 KQ435720 KOX78657.1 ODYU01000613 SOQ35703.1 ADTU01021133 MG573184 AWJ25046.1 DS235870 EEB19473.1 KQ976738 KYM75897.1 GBYB01006465 JAG76232.1 ACPB03000870 KQ977372 KYN03238.1 PZC79018.1 GDHC01011928 JAQ06701.1 ADTU01024445 ADTU01024446 ADTU01024447 ADTU01024448 ADTU01024449 ADTU01024450 ADTU01024451 ADTU01024452 GDQN01007992 GDQN01000519 JAT83062.1 JAT90535.1 KPJ09742.1 GEZM01041641 JAV80245.1 NNAY01000641 OXU27242.1 GECZ01031419 GECZ01031175 GECZ01028076 JAS38350.1 JAS38594.1 JAS41693.1 KPI97806.1 CM000070 KRT00510.1 KZ288222 PBC32081.1 JR047921 AEY60541.1 GFDG01000482 JAV18317.1 GBYB01013095 JAG82862.1 CH964272 EDW84092.1 CH902617 KPU80229.1 GAMC01015951 JAB90604.1 GAKP01017051 JAC41901.1 CH479179 EDW24136.1 KK107085 QOIP01000008 EZA60005.1 RLU19769.1 GECZ01022288 JAS47481.1 EAL28161.4 GDHF01023098 JAI29216.1 GBXI01017108 GBXI01014026 GBXI01003791 GBXI01000224 JAC97183.1 JAD00266.1 JAD10501.1 JAD14068.1 GEZM01041642 JAV80244.1 KK854125 PTY11739.1 EDV43365.1 KRT00511.1 GECL01001006 JAP05118.1 GAKP01017057 GAKP01017054 GAKP01017052 JAC41898.1 OWR46144.1 GAKP01017053 JAC41899.1 KPU80280.1 GFDF01002638 JAV11446.1 GAMC01015954 GAMC01015952 JAB90603.1 GGMS01014134 MBY83337.1 OUUW01000013 SPP87834.1 GAKP01014169 JAC44783.1 GDHF01010129 JAI42185.1 GBHO01033985 GDHC01002901 JAG09619.1 JAQ15728.1 GAKP01014168 JAC44784.1
Proteomes
UP000283053
UP000053240
UP000053268
UP000007151
UP000005204
UP000002358
+ More
UP000215335 UP000007266 UP000192223 UP000036403 UP000235965 UP000078492 UP000078541 UP000075809 UP000027135 UP000007755 UP000053825 UP000008237 UP000053105 UP000005205 UP000009046 UP000078540 UP000015103 UP000078542 UP000095301 UP000095300 UP000001819 UP000242457 UP000007798 UP000007801 UP000076408 UP000079169 UP000008744 UP000053097 UP000279307 UP000192221 UP000075900 UP000075884 UP000268350
UP000215335 UP000007266 UP000192223 UP000036403 UP000235965 UP000078492 UP000078541 UP000075809 UP000027135 UP000007755 UP000053825 UP000008237 UP000053105 UP000005205 UP000009046 UP000078540 UP000015103 UP000078542 UP000095301 UP000095300 UP000001819 UP000242457 UP000007798 UP000007801 UP000076408 UP000079169 UP000008744 UP000053097 UP000279307 UP000192221 UP000075900 UP000075884 UP000268350
PRIDE
ProteinModelPortal
A0A2H1V9U8
A0A3S2LQ19
A0A194QXQ3
A0A194PYI3
A0A2W1C1M5
A0A212EXC4
+ More
H9ISR4 I4DP69 A0A1E1W2F2 A0A1B6FP30 A0A1B6I0V4 A0A069DYP9 A0A290Y1Y8 A0A1B6EKQ3 A0A0T6BBD3 K7J7Y5 A0A232FGG0 A0A224XJ95 D6X366 A0A1W4WQD4 A0A0V0GA67 A0A0J7KVR6 J3JXW1 A0A2J7QVS3 A0A195EDU0 A0A195FWF1 A0A3S2TCH4 A0A151XA48 A0A1B6CS52 A0A067R7D7 A0A310S8Q7 A0A1B6KPQ9 A0A1Y1L5C0 A0A3Q8FM39 F4WLM2 A0A0L7RDI8 E2C8Q7 A0A0M9A788 A0A2H1V4D9 A0A158NN79 A0A3Q8G639 E0W1G7 A0A195AV15 A0A0C9RDQ9 T1HZW8 A0A195CR72 A0A2W1BVD6 A0A146LEX3 A0A158NS34 K7J7Y3 A0A1I8M815 A0A1E1W7T1 A0A194QW86 A0A1Y1M7G3 A0A232F9G3 A0A1B6EK88 A0A194PWR4 A0A1I8PE65 A0A0R3NHN9 A0A2A3EM15 V9IJ65 A0A1L8EHX7 A0A0C9RVW1 B4NKN1 A0A0P8YFQ5 W8BLU2 A0A034VF14 A0A1I8P5I0 A0A182YJR1 A0A1I8PDW3 A0A1S3D285 B4G2U1 A0A026WVC6 A0A1B6FBI1 Q297P4 A0A1I8MHJ2 A0A1W4VLW0 A0A182RI52 A0A0K8URN1 A0A0A1XHP0 A0A1Y1M310 A0A2R7VWM8 B3M241 A0A0R3NLI7 A0A182NCS1 A0A0V0GAQ6 A0A034VK19 A0A212EXH7 A0A034VHB6 A0A0P8YNC0 A0A1L8DYF1 W8B0N6 A0A2S2QZZ1 A0A3B0K0S6 A0A1I8P5B7 A0A034VTC3 A0A1W4VZ58 A0A0K8VUG2 A0A0A9WPG5 A0A034VQQ5
H9ISR4 I4DP69 A0A1E1W2F2 A0A1B6FP30 A0A1B6I0V4 A0A069DYP9 A0A290Y1Y8 A0A1B6EKQ3 A0A0T6BBD3 K7J7Y5 A0A232FGG0 A0A224XJ95 D6X366 A0A1W4WQD4 A0A0V0GA67 A0A0J7KVR6 J3JXW1 A0A2J7QVS3 A0A195EDU0 A0A195FWF1 A0A3S2TCH4 A0A151XA48 A0A1B6CS52 A0A067R7D7 A0A310S8Q7 A0A1B6KPQ9 A0A1Y1L5C0 A0A3Q8FM39 F4WLM2 A0A0L7RDI8 E2C8Q7 A0A0M9A788 A0A2H1V4D9 A0A158NN79 A0A3Q8G639 E0W1G7 A0A195AV15 A0A0C9RDQ9 T1HZW8 A0A195CR72 A0A2W1BVD6 A0A146LEX3 A0A158NS34 K7J7Y3 A0A1I8M815 A0A1E1W7T1 A0A194QW86 A0A1Y1M7G3 A0A232F9G3 A0A1B6EK88 A0A194PWR4 A0A1I8PE65 A0A0R3NHN9 A0A2A3EM15 V9IJ65 A0A1L8EHX7 A0A0C9RVW1 B4NKN1 A0A0P8YFQ5 W8BLU2 A0A034VF14 A0A1I8P5I0 A0A182YJR1 A0A1I8PDW3 A0A1S3D285 B4G2U1 A0A026WVC6 A0A1B6FBI1 Q297P4 A0A1I8MHJ2 A0A1W4VLW0 A0A182RI52 A0A0K8URN1 A0A0A1XHP0 A0A1Y1M310 A0A2R7VWM8 B3M241 A0A0R3NLI7 A0A182NCS1 A0A0V0GAQ6 A0A034VK19 A0A212EXH7 A0A034VHB6 A0A0P8YNC0 A0A1L8DYF1 W8B0N6 A0A2S2QZZ1 A0A3B0K0S6 A0A1I8P5B7 A0A034VTC3 A0A1W4VZ58 A0A0K8VUG2 A0A0A9WPG5 A0A034VQQ5
Ontologies
KEGG
GO
PANTHER
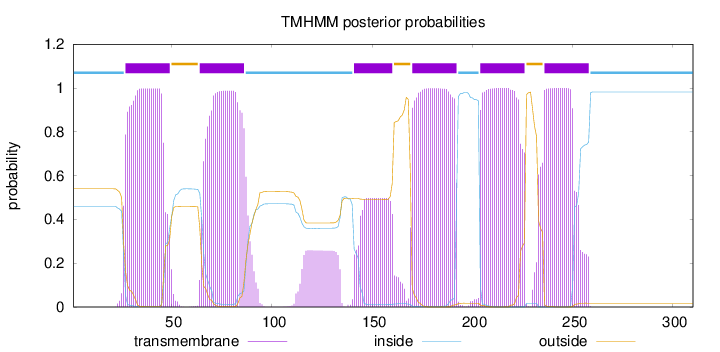
Topology
Length:
310
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
122.6786
Exp number, first 60 AAs:
21.16135
Total prob of N-in:
0.46009
POSSIBLE N-term signal
sequence
inside
1 - 26
TMhelix
27 - 49
outside
50 - 63
TMhelix
64 - 86
inside
87 - 140
TMhelix
141 - 160
outside
161 - 169
TMhelix
170 - 192
inside
193 - 203
TMhelix
204 - 226
outside
227 - 235
TMhelix
236 - 258
inside
259 - 310
Population Genetic Test Statistics
Pi
220.298855
Theta
173.256657
Tajima's D
0.826355
CLR
0.11597
CSRT
0.606819659017049
Interpretation
Uncertain
