Gene
KWMTBOMO13033
Pre Gene Modal
BGIBMGA000422
Annotation
elongation_of_very_long_chain_fatty_acids_protein_AAEL008004_[Papilio_xuthus]
Full name
Elongation of very long chain fatty acids protein
Alternative Name
Very-long-chain 3-oxoacyl-CoA synthase
Location in the cell
PlasmaMembrane Reliability : 4.722
Sequence
CDS
ATGGACAGCAGGGGCGATCCTCGGGTGAAAGACTGGCCGATGATGTCATCGCCGTGGCCAACCCTAGCCGCGTGCGTGTGCTACGCATACTGCGCGAAGGAGCTTGGCCCCAAACTTATGGCCAATCGAAAACCATTCGAACTTCGAAATATTCTCGTCGTTTACAACATGGCTCAGACTATCTTCAGCGCCTGGATATTCTACGAGTATTTGATGAGCGGATGGTGGGGCCACTATGACTTTAGGTGTCAACTGGTAGATTACTCTCGGAGTCCAATGGCCATGAGGATGGCGAACACGTGTTGGTGGTACTACTTCAGCAAGTTCACAGAATTTATGGACACGCTTTTCTTTGTGCTACGGAAGAAGAACGAGCACGTGTCCACGTTGCATGTGATACATCACGGGATCATGCCGATGTCTGTTTGGTTCGGACTTAAATTCGCGCCGGGTGGTCACAGCACTTTCTTCGCTCTGCTCAACACGTTCGTTCACATCGTAATGTACTTTTACTACATGGTGGCTGCCATGGGTCCTAAGTATCAGAAGTATATCTGGTGGAAGAAATACCTCACAGCCTTCCAGATGGTTCAGTTCGTATTGATCTTTAGTCATCAACTCCAAGTCCTCTTCAGGCCGTCGTGTCAATACCCTCGTGTCTTCGTCTACTGGATCGCGATGCACGGCTTCTTATTCTTGTTCCTCTTCAGCGATTTCTACAAAGCGAGGTACACAACCGCCGAGAGGAAATCGAAGGCTCAGAACGGACTTTGTATGGCGGTGATGGACGACAGCTCGAGTTCGCTGAACGGCAGGAACGGCTACAAGCAGGAGATGGGGTCGGAGGTGCCCAGTTCGTACGCCTCGTCCGGGGCGGACGCGTTCGTGCGTCGGAGACCGCTCTCATAG
Protein
MDSRGDPRVKDWPMMSSPWPTLAACVCYAYCAKELGPKLMANRKPFELRNILVVYNMAQTIFSAWIFYEYLMSGWWGHYDFRCQLVDYSRSPMAMRMANTCWWYYFSKFTEFMDTLFFVLRKKNEHVSTLHVIHHGIMPMSVWFGLKFAPGGHSTFFALLNTFVHIVMYFYYMVAAMGPKYQKYIWWKKYLTAFQMVQFVLIFSHQLQVLFRPSCQYPRVFVYWIAMHGFLFLFLFSDFYKARYTTAERKSKAQNGLCMAVMDDSSSSLNGRNGYKQEMGSEVPSSYASSGADAFVRRRPLS
Summary
Catalytic Activity
a very-long-chain acyl-CoA + H(+) + malonyl-CoA = a very-long-chain 3-oxoacyl-CoA + CO2 + CoA
Similarity
Belongs to the ELO family.
Uniprot
H9IT43
A0A2W1BVK9
B2DBH6
A0A194R1L2
S4P8U6
A0A1E1WBM2
+ More
A0A212EXD6 A0A224XG77 A0A0V0G986 A0A023F5B4 A0A069DSG8 A0A0P4VQ12 A0A1B6D8X5 A0A290Y213 A0A1B6INS9 A0A1B6L1U0 D6X365 A0A1B6FFP4 A0A0K8TP99 A0A1B6GMU5 A0A1I8M7X8 A0A0L7RD69 A0A3Q8FM02 A0A0K8VMK5 A0A088AIA2 B3M2T7 A0A034VI44 B4M0X5 A0A1L8DY78 A0A3B0KWA1 A0A026WWA2 B4I3P6 B4KA49 A0A1B0A8T5 B3P216 J9K2C4 E1ZZH1 A0A336KQL3 A0A195EEV2 A0A2H8TI43 Q295H0 B4PTT6 A0A0A1WVQ5 A0A195AV43 A0A195FWS7 B4QVI8 Q95T98 A0A1W4VXQ5 A0A0R3NP99 A0A195CSY9 B4GDW9 A0A1Y1MLN6 A0A2A3ELP3 B4NIB7 B4JI95 A0A1A9W6N2 F4WLM1 A0A182PLE3 Q16VG4 A0A1S4FMR5 A0A0A9Z4G4 A0A067RGR0 A0A182K8R4 A0A182W353 A0A182NCS0 U5ETZ3 A0A182QFM6 A0A182XF38 A0A151XA50 A0A182J6Z7 W8AXR8 A0A1L8EIJ4 A0A182HVK6 A0A182RI53 Q7PHE2 A0A232FGZ5 A0A1Q3G0K7 A0A1Q3G0L3 A0A2M3Z940 A0A1Q3G0M3 A0A310SHJ3 A0A2M4CMQ7 A0A2M4CMW4 A0A2M4AQW2 K7J7Y4 A0A1I8P686 W5JG42 A0A0M4EJT0 A0A2P8Z876 A0A182KZX1 A0A0K8TMV7 A0A034VHB6 E2C8Q5 A0A182YJR0 A0A0K8URN1 A0A0K8V5J5 A0A1W4WQD4 A0A034VHC0 A0A1L8EI23 A0A1I8P5D4
A0A212EXD6 A0A224XG77 A0A0V0G986 A0A023F5B4 A0A069DSG8 A0A0P4VQ12 A0A1B6D8X5 A0A290Y213 A0A1B6INS9 A0A1B6L1U0 D6X365 A0A1B6FFP4 A0A0K8TP99 A0A1B6GMU5 A0A1I8M7X8 A0A0L7RD69 A0A3Q8FM02 A0A0K8VMK5 A0A088AIA2 B3M2T7 A0A034VI44 B4M0X5 A0A1L8DY78 A0A3B0KWA1 A0A026WWA2 B4I3P6 B4KA49 A0A1B0A8T5 B3P216 J9K2C4 E1ZZH1 A0A336KQL3 A0A195EEV2 A0A2H8TI43 Q295H0 B4PTT6 A0A0A1WVQ5 A0A195AV43 A0A195FWS7 B4QVI8 Q95T98 A0A1W4VXQ5 A0A0R3NP99 A0A195CSY9 B4GDW9 A0A1Y1MLN6 A0A2A3ELP3 B4NIB7 B4JI95 A0A1A9W6N2 F4WLM1 A0A182PLE3 Q16VG4 A0A1S4FMR5 A0A0A9Z4G4 A0A067RGR0 A0A182K8R4 A0A182W353 A0A182NCS0 U5ETZ3 A0A182QFM6 A0A182XF38 A0A151XA50 A0A182J6Z7 W8AXR8 A0A1L8EIJ4 A0A182HVK6 A0A182RI53 Q7PHE2 A0A232FGZ5 A0A1Q3G0K7 A0A1Q3G0L3 A0A2M3Z940 A0A1Q3G0M3 A0A310SHJ3 A0A2M4CMQ7 A0A2M4CMW4 A0A2M4AQW2 K7J7Y4 A0A1I8P686 W5JG42 A0A0M4EJT0 A0A2P8Z876 A0A182KZX1 A0A0K8TMV7 A0A034VHB6 E2C8Q5 A0A182YJR0 A0A0K8URN1 A0A0K8V5J5 A0A1W4WQD4 A0A034VHC0 A0A1L8EI23 A0A1I8P5D4
EC Number
2.3.1.199
Pubmed
19121390
28756777
18712529
26354079
23622113
22118469
+ More
25474469 26334808 27129103 28887463 18362917 19820115 26369729 25315136 17994087 25348373 18057021 24508170 30249741 20798317 15632085 23185243 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 21719571 17510324 25401762 26823975 24845553 24495485 12364791 14747013 17210077 28648823 20075255 20920257 23761445 29403074 20966253 25244985
25474469 26334808 27129103 28887463 18362917 19820115 26369729 25315136 17994087 25348373 18057021 24508170 30249741 20798317 15632085 23185243 17550304 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 28004739 21719571 17510324 25401762 26823975 24845553 24495485 12364791 14747013 17210077 28648823 20075255 20920257 23761445 29403074 20966253 25244985
EMBL
BABH01013313
KZ149892
PZC79019.1
AB264656
KQ459586
BAG30735.1
+ More
KPI97808.1 KQ461073 KPJ09741.1 GAIX01007127 JAA85433.1 GDQN01009681 GDQN01006773 GDQN01003504 JAT81373.1 JAT84281.1 JAT87550.1 AGBW02011787 OWR46145.1 GFTR01004941 JAW11485.1 GECL01001796 JAP04328.1 GBBI01002413 JAC16299.1 GBGD01002208 JAC86681.1 GDKW01002097 JAI54498.1 GEDC01020363 GEDC01015159 GEDC01006393 GEDC01005729 JAS16935.1 JAS22139.1 JAS30905.1 JAS31569.1 MF279188 ATD85028.1 GECU01019121 JAS88585.1 GEBQ01022473 JAT17504.1 KQ971372 EFA09796.1 GECZ01020764 JAS49005.1 GDAI01001632 JAI15971.1 GECZ01006010 JAS63759.1 KQ414614 KOC68912.1 MG573190 AWJ25052.1 GDHF01027266 GDHF01014458 GDHF01013658 GDHF01012192 GDHF01005627 JAI25048.1 JAI37856.1 JAI38656.1 JAI40122.1 JAI46687.1 CH902617 EDV43467.1 GAKP01017160 GAKP01017158 GAKP01017154 GAKP01017151 JAC41792.1 CH940650 EDW67386.1 KRF83276.1 GFDF01002667 JAV11417.1 OUUW01000014 SPP88268.1 KK107085 QOIP01000008 EZA60006.1 RLU19768.1 CH480821 EDW54839.1 CH933806 EDW16724.1 CH954181 EDV47789.1 ABLF02039120 ABLF02039121 GL435347 EFN73422.1 UFQS01000831 UFQT01000831 SSX07116.1 SSX27459.1 KQ979039 KYN23389.1 GFXV01001990 MBW13795.1 CM000070 EAL28742.1 CM000160 EDW95669.1 GBXI01011591 JAD02701.1 KQ976738 KYM75895.1 KQ981208 KYN44772.1 CM000364 EDX11613.1 AY060267 AE014297 AAL25306.1 AAN13323.1 KRT01005.1 KQ977372 KYN03239.1 CH479182 EDW34630.1 GEZM01027911 JAV86583.1 KZ288222 PBC32079.1 CH964272 EDW83699.1 CH916369 EDV92976.1 GL888208 EGI64957.1 CH477593 EAT38554.1 GBHO01035038 GBHO01003487 GBHO01003486 GBHO01003483 GBHO01003482 GBHO01003481 GBRD01015936 GBRD01015934 GDHC01018821 GDHC01008178 JAG08566.1 JAG40117.1 JAG40118.1 JAG40121.1 JAG40122.1 JAG40123.1 JAG49890.1 JAP99807.1 JAQ10451.1 KK852651 KDR19452.1 GANO01002561 JAB57310.1 AXCN02001584 KQ982351 KYQ57246.1 GAMC01012775 GAMC01012774 JAB93780.1 GFDG01000353 JAV18446.1 APCN01005798 AAAB01008898 EAA44575.5 NNAY01000231 OXU29833.1 GFDL01001710 JAV33335.1 GFDL01001715 JAV33330.1 GGFM01004254 MBW25005.1 GFDL01001713 JAV33332.1 KQ768821 OAD53050.1 GGFL01002442 MBW66620.1 GGFL01002441 MBW66619.1 GGFK01009853 MBW43174.1 ADMH02001497 ETN62308.1 CP012526 ALC45265.1 ALC47358.1 PYGN01000151 PSN52697.1 GDAI01001911 JAI15692.1 GAKP01017053 JAC41899.1 GL453671 EFN75679.1 GDHF01023098 JAI29216.1 GDHF01018117 JAI34197.1 GAKP01017058 GAKP01017055 GAKP01017048 JAC41904.1 GFDG01000481 JAV18318.1
KPI97808.1 KQ461073 KPJ09741.1 GAIX01007127 JAA85433.1 GDQN01009681 GDQN01006773 GDQN01003504 JAT81373.1 JAT84281.1 JAT87550.1 AGBW02011787 OWR46145.1 GFTR01004941 JAW11485.1 GECL01001796 JAP04328.1 GBBI01002413 JAC16299.1 GBGD01002208 JAC86681.1 GDKW01002097 JAI54498.1 GEDC01020363 GEDC01015159 GEDC01006393 GEDC01005729 JAS16935.1 JAS22139.1 JAS30905.1 JAS31569.1 MF279188 ATD85028.1 GECU01019121 JAS88585.1 GEBQ01022473 JAT17504.1 KQ971372 EFA09796.1 GECZ01020764 JAS49005.1 GDAI01001632 JAI15971.1 GECZ01006010 JAS63759.1 KQ414614 KOC68912.1 MG573190 AWJ25052.1 GDHF01027266 GDHF01014458 GDHF01013658 GDHF01012192 GDHF01005627 JAI25048.1 JAI37856.1 JAI38656.1 JAI40122.1 JAI46687.1 CH902617 EDV43467.1 GAKP01017160 GAKP01017158 GAKP01017154 GAKP01017151 JAC41792.1 CH940650 EDW67386.1 KRF83276.1 GFDF01002667 JAV11417.1 OUUW01000014 SPP88268.1 KK107085 QOIP01000008 EZA60006.1 RLU19768.1 CH480821 EDW54839.1 CH933806 EDW16724.1 CH954181 EDV47789.1 ABLF02039120 ABLF02039121 GL435347 EFN73422.1 UFQS01000831 UFQT01000831 SSX07116.1 SSX27459.1 KQ979039 KYN23389.1 GFXV01001990 MBW13795.1 CM000070 EAL28742.1 CM000160 EDW95669.1 GBXI01011591 JAD02701.1 KQ976738 KYM75895.1 KQ981208 KYN44772.1 CM000364 EDX11613.1 AY060267 AE014297 AAL25306.1 AAN13323.1 KRT01005.1 KQ977372 KYN03239.1 CH479182 EDW34630.1 GEZM01027911 JAV86583.1 KZ288222 PBC32079.1 CH964272 EDW83699.1 CH916369 EDV92976.1 GL888208 EGI64957.1 CH477593 EAT38554.1 GBHO01035038 GBHO01003487 GBHO01003486 GBHO01003483 GBHO01003482 GBHO01003481 GBRD01015936 GBRD01015934 GDHC01018821 GDHC01008178 JAG08566.1 JAG40117.1 JAG40118.1 JAG40121.1 JAG40122.1 JAG40123.1 JAG49890.1 JAP99807.1 JAQ10451.1 KK852651 KDR19452.1 GANO01002561 JAB57310.1 AXCN02001584 KQ982351 KYQ57246.1 GAMC01012775 GAMC01012774 JAB93780.1 GFDG01000353 JAV18446.1 APCN01005798 AAAB01008898 EAA44575.5 NNAY01000231 OXU29833.1 GFDL01001710 JAV33335.1 GFDL01001715 JAV33330.1 GGFM01004254 MBW25005.1 GFDL01001713 JAV33332.1 KQ768821 OAD53050.1 GGFL01002442 MBW66620.1 GGFL01002441 MBW66619.1 GGFK01009853 MBW43174.1 ADMH02001497 ETN62308.1 CP012526 ALC45265.1 ALC47358.1 PYGN01000151 PSN52697.1 GDAI01001911 JAI15692.1 GAKP01017053 JAC41899.1 GL453671 EFN75679.1 GDHF01023098 JAI29216.1 GDHF01018117 JAI34197.1 GAKP01017058 GAKP01017055 GAKP01017048 JAC41904.1 GFDG01000481 JAV18318.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000007151
UP000007266
UP000095301
+ More
UP000053825 UP000005203 UP000007801 UP000008792 UP000268350 UP000053097 UP000279307 UP000001292 UP000009192 UP000092445 UP000008711 UP000007819 UP000000311 UP000078492 UP000001819 UP000002282 UP000078540 UP000078541 UP000000304 UP000000803 UP000192221 UP000078542 UP000008744 UP000242457 UP000007798 UP000001070 UP000091820 UP000007755 UP000075885 UP000008820 UP000027135 UP000075881 UP000075920 UP000075884 UP000075886 UP000076407 UP000075809 UP000075880 UP000075840 UP000075900 UP000007062 UP000215335 UP000002358 UP000095300 UP000000673 UP000092553 UP000245037 UP000075882 UP000008237 UP000076408 UP000192223
UP000053825 UP000005203 UP000007801 UP000008792 UP000268350 UP000053097 UP000279307 UP000001292 UP000009192 UP000092445 UP000008711 UP000007819 UP000000311 UP000078492 UP000001819 UP000002282 UP000078540 UP000078541 UP000000304 UP000000803 UP000192221 UP000078542 UP000008744 UP000242457 UP000007798 UP000001070 UP000091820 UP000007755 UP000075885 UP000008820 UP000027135 UP000075881 UP000075920 UP000075884 UP000075886 UP000076407 UP000075809 UP000075880 UP000075840 UP000075900 UP000007062 UP000215335 UP000002358 UP000095300 UP000000673 UP000092553 UP000245037 UP000075882 UP000008237 UP000076408 UP000192223
Pfam
PF01151 ELO
Interpro
IPR002076
ELO_fam
ProteinModelPortal
H9IT43
A0A2W1BVK9
B2DBH6
A0A194R1L2
S4P8U6
A0A1E1WBM2
+ More
A0A212EXD6 A0A224XG77 A0A0V0G986 A0A023F5B4 A0A069DSG8 A0A0P4VQ12 A0A1B6D8X5 A0A290Y213 A0A1B6INS9 A0A1B6L1U0 D6X365 A0A1B6FFP4 A0A0K8TP99 A0A1B6GMU5 A0A1I8M7X8 A0A0L7RD69 A0A3Q8FM02 A0A0K8VMK5 A0A088AIA2 B3M2T7 A0A034VI44 B4M0X5 A0A1L8DY78 A0A3B0KWA1 A0A026WWA2 B4I3P6 B4KA49 A0A1B0A8T5 B3P216 J9K2C4 E1ZZH1 A0A336KQL3 A0A195EEV2 A0A2H8TI43 Q295H0 B4PTT6 A0A0A1WVQ5 A0A195AV43 A0A195FWS7 B4QVI8 Q95T98 A0A1W4VXQ5 A0A0R3NP99 A0A195CSY9 B4GDW9 A0A1Y1MLN6 A0A2A3ELP3 B4NIB7 B4JI95 A0A1A9W6N2 F4WLM1 A0A182PLE3 Q16VG4 A0A1S4FMR5 A0A0A9Z4G4 A0A067RGR0 A0A182K8R4 A0A182W353 A0A182NCS0 U5ETZ3 A0A182QFM6 A0A182XF38 A0A151XA50 A0A182J6Z7 W8AXR8 A0A1L8EIJ4 A0A182HVK6 A0A182RI53 Q7PHE2 A0A232FGZ5 A0A1Q3G0K7 A0A1Q3G0L3 A0A2M3Z940 A0A1Q3G0M3 A0A310SHJ3 A0A2M4CMQ7 A0A2M4CMW4 A0A2M4AQW2 K7J7Y4 A0A1I8P686 W5JG42 A0A0M4EJT0 A0A2P8Z876 A0A182KZX1 A0A0K8TMV7 A0A034VHB6 E2C8Q5 A0A182YJR0 A0A0K8URN1 A0A0K8V5J5 A0A1W4WQD4 A0A034VHC0 A0A1L8EI23 A0A1I8P5D4
A0A212EXD6 A0A224XG77 A0A0V0G986 A0A023F5B4 A0A069DSG8 A0A0P4VQ12 A0A1B6D8X5 A0A290Y213 A0A1B6INS9 A0A1B6L1U0 D6X365 A0A1B6FFP4 A0A0K8TP99 A0A1B6GMU5 A0A1I8M7X8 A0A0L7RD69 A0A3Q8FM02 A0A0K8VMK5 A0A088AIA2 B3M2T7 A0A034VI44 B4M0X5 A0A1L8DY78 A0A3B0KWA1 A0A026WWA2 B4I3P6 B4KA49 A0A1B0A8T5 B3P216 J9K2C4 E1ZZH1 A0A336KQL3 A0A195EEV2 A0A2H8TI43 Q295H0 B4PTT6 A0A0A1WVQ5 A0A195AV43 A0A195FWS7 B4QVI8 Q95T98 A0A1W4VXQ5 A0A0R3NP99 A0A195CSY9 B4GDW9 A0A1Y1MLN6 A0A2A3ELP3 B4NIB7 B4JI95 A0A1A9W6N2 F4WLM1 A0A182PLE3 Q16VG4 A0A1S4FMR5 A0A0A9Z4G4 A0A067RGR0 A0A182K8R4 A0A182W353 A0A182NCS0 U5ETZ3 A0A182QFM6 A0A182XF38 A0A151XA50 A0A182J6Z7 W8AXR8 A0A1L8EIJ4 A0A182HVK6 A0A182RI53 Q7PHE2 A0A232FGZ5 A0A1Q3G0K7 A0A1Q3G0L3 A0A2M3Z940 A0A1Q3G0M3 A0A310SHJ3 A0A2M4CMQ7 A0A2M4CMW4 A0A2M4AQW2 K7J7Y4 A0A1I8P686 W5JG42 A0A0M4EJT0 A0A2P8Z876 A0A182KZX1 A0A0K8TMV7 A0A034VHB6 E2C8Q5 A0A182YJR0 A0A0K8URN1 A0A0K8V5J5 A0A1W4WQD4 A0A034VHC0 A0A1L8EI23 A0A1I8P5D4
Ontologies
PATHWAY
GO
PANTHER
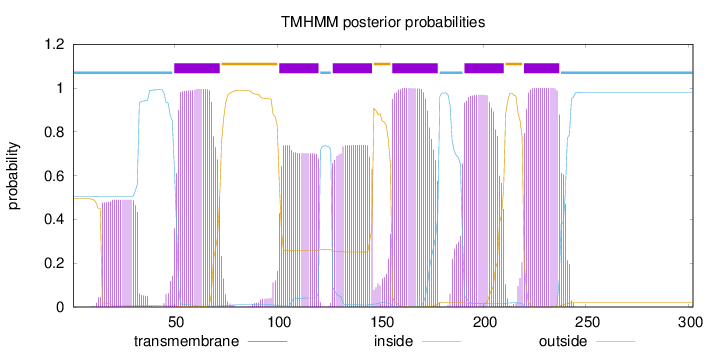
Topology
Length:
302
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
121.70714
Exp number, first 60 AAs:
19.23419
Total prob of N-in:
0.50556
POSSIBLE N-term signal
sequence
inside
1 - 49
TMhelix
50 - 72
outside
73 - 100
TMhelix
101 - 120
inside
121 - 126
TMhelix
127 - 146
outside
147 - 155
TMhelix
156 - 178
inside
179 - 190
TMhelix
191 - 210
outside
211 - 219
TMhelix
220 - 237
inside
238 - 302
Population Genetic Test Statistics
Pi
210.095787
Theta
171.965441
Tajima's D
0.614918
CLR
0.237256
CSRT
0.552022398880056
Interpretation
Uncertain
