Gene
KWMTBOMO13008 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000273
Annotation
cuticular_protein_RR-2_motif_91_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.796
Sequence
CDS
ATGATCGCTAAAGCCGCAGTAATCCTGTCCGTTGTAGCACTAGCAAGTGCTGGTGTAGTCCAACTAGCAGGCCACGGATACGGCGGCGAGGAATACGGCCACGAAGCGATCGCGTACGCCCCAGTAGCCGTTGCGCCAGTCGCGCATTACGCCGCACACGAAGACACGCATGTCGATTATCATGCTCATCCCAAATATGACTACTCCTACTCAGTGTCAGACCCTCATACCGGCGACCATAAGACCCAGCATGAGGCCCGCGATGGTGATGTCGTAAAGGGAGAGTACTCTTTGCTCCAGCCCGACGGCTCCTTCAGAAAAGTTACATACACTGCTGACGACCATAATGGTTTCAACGCAGTAGTACACAACACCGCACCCGCTCATCACGAGTACCACCACTAG
Protein
MIAKAAVILSVVALASAGVVQLAGHGYGGEEYGHEAIAYAPVAVAPVAHYAAHEDTHVDYHAHPKYDYSYSVSDPHTGDHKTQHEARDGDVVKGEYSLLQPDGSFRKVTYTADDHNGFNAVVHNTAPAHHEYHH
Summary
Uniprot
ProteinModelPortal
Ontologies
GO
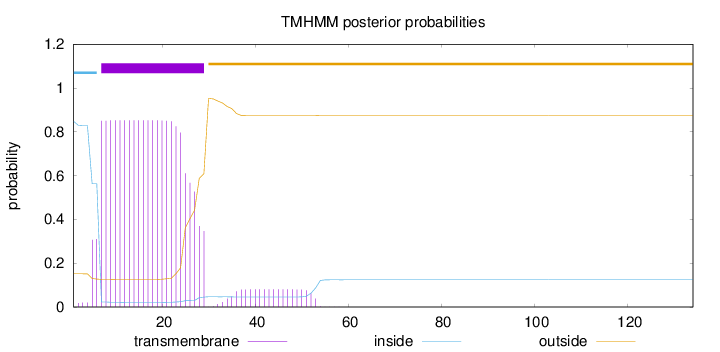
Topology
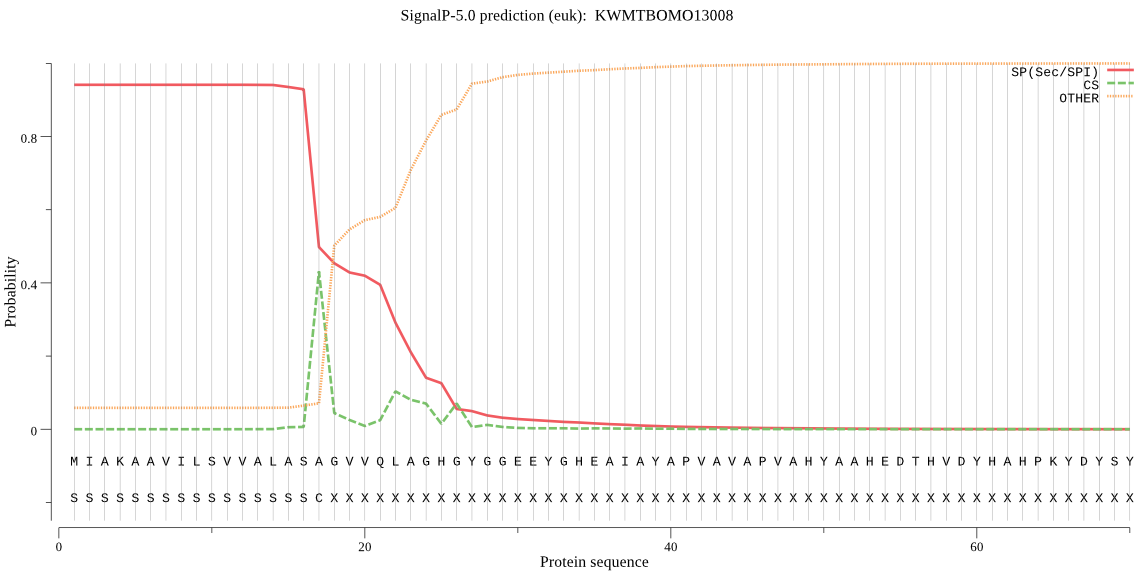
SignalP
Position: 1 - 17,
Likelihood: 0.941394
Length:
134
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.8444
Exp number, first 60 AAs:
19.84439
Total prob of N-in:
0.84850
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 134
Population Genetic Test Statistics
Pi
244.59101
Theta
207.231146
Tajima's D
0.678828
CLR
0.020538
CSRT
0.568321583920804
Interpretation
Uncertain
