Gene
KWMTBOMO13005 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000270
Annotation
cuticular_protein_RR-2_motif_94_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.064
Sequence
CDS
ATGTTTTCCAAATTAATAGTACTGTGCGCATTTGTGGCGGTGTCCAAAGCTGGTCTCCTGGCGGAACCAGTCCACTACTCATCTGCTGGGGCAGTTTCCTCCCAGAGCATCGTACGCCATGACGAAAGCCACGGTGCTCATTATGCCGCTGCGGCTCCTGTAGCTTACCACGCTGCCCCTGTGGCTTACCATGCTGCCCCTGTAGCGTACCATGCTGCTCCAGTCCACTATTCGTCTGCCGAGTCCGTCTCTTCCCAGAACATCATCCGTCATGATCAGCCCCAGACTATCAATTACGCTGCTCCCGTAGCCAAGTTAGCCGTCGCTACACCCGCCTACCCTAAATACGAGTACAACTACTCAGTAGCTGACGGTCACTCCGGCGACAACAAGTCCCAACAAGAAGTCCGCGACGGTGACGCAGTGAAGGGCTCATACTCCTTCCACGAAGCTGACGGCTCCATCAGAACCGTTGAGTACACCGCTGATGCACACAACGGCTTCAACGCTGTCGTCCACAACACCGCCCCCACCGCCGCCCCCACACACATCAAGGCTAGATATCCGAAATATGAATTCGAATATTCAGTGTCAGACAAAAAGACTGGGGACCACAAAACTCATCGTGAGTCGCGTGACGGAGATCGTGTTCGTGGTGAATACAGCCTTGTCGAACCTGATGGTTCGCTGCGGAAGGTGGAATACGATGCTGATGACCATAACGGTTTCAGCGCTGTTGTAAGTAAAGGTGTACATAAGCACGGTGACCACGCATACTCAGTTTTTGGTCACACCAGACACTTTTACCCTATCGGACATGGCATCAAGATCAATCACTATTTCCCAGAAAAGAACACCGCAAAAGACGTTAGCTTTGATAACACTGATAAAGGAAAGCCTGTTCAAGAAAATGTACAGGATTCCGTTGGTTTTCAAAAGAGTGCCACTAAGACTGATGTTGAAACATCAGATAAAGAAATTACTTTAAAACCAGAAAATGCTGAAAATGTTGGAAAATCAAATGCCGAGAGACTTATTCAAAAGGATTTAGTCGCAGTGGTCGATTCGACAACTCCTTTGATTATTGAAAATGAGAAATCACTTGAAGGAAGCACAGAAGAGATCAATACTGTAACAACTGAAACTTCCGTAATTGATGCTATGAAAGGGGAAGAGATAATAAAAAAAGATGACCCCAGTCCCGACAACGAAGTAGCTTCGTCACACTACCATATGTTTTATTACGTTTATTGA
Protein
MFSKLIVLCAFVAVSKAGLLAEPVHYSSAGAVSSQSIVRHDESHGAHYAAAAPVAYHAAPVAYHAAPVAYHAAPVHYSSAESVSSQNIIRHDQPQTINYAAPVAKLAVATPAYPKYEYNYSVADGHSGDNKSQQEVRDGDAVKGSYSFHEADGSIRTVEYTADAHNGFNAVVHNTAPTAAPTHIKARYPKYEFEYSVSDKKTGDHKTHRESRDGDRVRGEYSLVEPDGSLRKVEYDADDHNGFSAVVSKGVHKHGDHAYSVFGHTRHFYPIGHGIKINHYFPEKNTAKDVSFDNTDKGKPVQENVQDSVGFQKSATKTDVETSDKEITLKPENAENVGKSNAERLIQKDLVAVVDSTTPLIIENEKSLEGSTEEINTVTTETSVIDAMKGEEIIKKDDPSPDNEVASSHYHMFYYVY
Summary
Uniprot
EMBL
Pfam
PF00379 Chitin_bind_4
ProteinModelPortal
Ontologies
GO
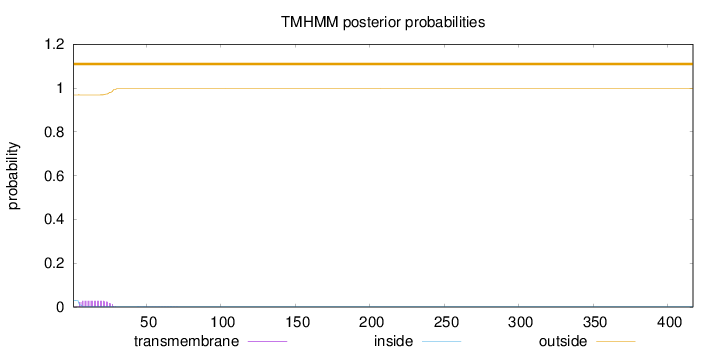
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.957921

Length:
417
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.621399999999999
Exp number, first 60 AAs:
0.61511
Total prob of N-in:
0.03138
outside
1 - 417
Population Genetic Test Statistics
Pi
248.353355
Theta
215.268392
Tajima's D
0.442124
CLR
1.379069
CSRT
0.489525523723814
Interpretation
Uncertain
