Gene
KWMTBOMO13000 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000264
Annotation
cuticular_protein_RR-2_motif_100_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.618 PlasmaMembrane Reliability : 1.06
Sequence
CDS
ATGATTTCCAAAATCGTTGTACTGTGTGCCTTTGTGGCAGTCTCGCAGGCCGGTCTCTTGGCTGCGCCCGTGCACTACTCTCCCGCTGAGGCTGTTTCCTCCCAAAGCATCGTACGTCATGATCAGGCTCACGGCACTAAGCTTGTAGCTCCCGTTGCCAAGCTCGCCATTGCCGCACCTGTTGCATACCACGCTGCTCCAGCTCCAATCGCCTACCACGCTGCCCCCGCTCCCATTGCCTACCATGCCCCCATCGCCAAAGTGCTAGCTCACCGTGAAGACCAAGAAATCGCTTATCCAAAATACGAGTACAACTACTCAGTAGCTGACGGTCACTCCGGCGACAACAAGTCCCAACAAGAAGTCCGCGATGGCGATGTTGTAAAGGGTTCATACTCCTTCCACGAAGCTGACGGCTCTATCAGAACTGTTGAGTACACCGCTGACGACCACAACGGTTTCAACGCGGTCGTCCACAACACTGCCCCTACCGCTGCCCCCACACACATCAAGGCTTTGCCTGCACTCCAATACTACCATCACTAA
Protein
MISKIVVLCAFVAVSQAGLLAAPVHYSPAEAVSSQSIVRHDQAHGTKLVAPVAKLAIAAPVAYHAAPAPIAYHAAPAPIAYHAPIAKVLAHREDQEIAYPKYEYNYSVADGHSGDNKSQQEVRDGDVVKGSYSFHEADGSIRTVEYTADDHNGFNAVVHNTAPTAAPTHIKALPALQYYHH
Summary
Uniprot
C0H6U2
H9ISN7
C0H6T8
C0H6T9
C0H6T7
Q9BPR6
+ More
C0H6U1 C0H6U0 Q9BPR7 B2DBJ3 A0A194PWP3 I4DRA3 I4DJP1 A0A212F1C1 A0A194QWT1 A0A194QXS8 A0A2W1BXD1 A0A2A4J2S9 I4DRA2 A0A212F1A8 A0A2W1BV71 A0A2W1BZV6 I4DKM4 A0A194Q302 B2DBK0 A0A2W1BVB0 A0A194PYF6 A0A2W1C485 A0A0N1IPH5 A0A212EJB4 A0A2H1W3Y2 A0A2A4J4D6 A0A2A4J4N1 A0A2H1WU56 A0A2H1VP46 A0A2W1BVH0 A0A2A4IXI8 A0A2W1C0G8 C0H6U5 A0A2H1VQS4 H9IT50 C0H6U4 C0H6U6 Q9BPR9 A0A2W1BZH0 Q9BPR8 I4DMR3 C0H6U3 C0H6U7 I4DK03 I4DMH3 C0H6S9 C0H6S8 A0A212F891 I4DJZ8 A0A194PWQ0 A0A194QW46 A0A194R1N5 A0A194PWP0 A0A212F8A9 A0A212F9Z8 A0A194QWS6
C0H6U1 C0H6U0 Q9BPR7 B2DBJ3 A0A194PWP3 I4DRA3 I4DJP1 A0A212F1C1 A0A194QWT1 A0A194QXS8 A0A2W1BXD1 A0A2A4J2S9 I4DRA2 A0A212F1A8 A0A2W1BV71 A0A2W1BZV6 I4DKM4 A0A194Q302 B2DBK0 A0A2W1BVB0 A0A194PYF6 A0A2W1C485 A0A0N1IPH5 A0A212EJB4 A0A2H1W3Y2 A0A2A4J4D6 A0A2A4J4N1 A0A2H1WU56 A0A2H1VP46 A0A2W1BVH0 A0A2A4IXI8 A0A2W1C0G8 C0H6U5 A0A2H1VQS4 H9IT50 C0H6U4 C0H6U6 Q9BPR9 A0A2W1BZH0 Q9BPR8 I4DMR3 C0H6U3 C0H6U7 I4DK03 I4DMH3 C0H6S9 C0H6S8 A0A212F891 I4DJZ8 A0A194PWQ0 A0A194QW46 A0A194R1N5 A0A194PWP0 A0A212F8A9 A0A212F9Z8 A0A194QWS6
EMBL
BR000601
FAA00603.1
BABH01013343
BR000597
FAA00599.1
BR000598
+ More
FAA00600.1 BR000596 FAA00598.1 AB047481 BAB32478.1 BR000600 FAA00602.1 BR000599 FAA00601.1 AB047480 BAB32477.1 AB264676 BAG30752.1 KQ459586 KPI97786.1 AK405036 BAM20443.1 AK401509 BAM18131.1 AGBW02010913 OWR47527.1 KQ461073 KPJ09764.1 KPJ09765.1 KZ149892 PZC78981.1 NWSH01003366 PCG66457.1 AK405035 BAM20442.1 OWR47528.1 PZC78982.1 PZC78985.1 AK401842 BAM18464.1 KPI97785.1 AB264683 BAG30759.1 PZC78988.1 KPI97784.1 PZC78983.1 KQ460333 KPJ15643.1 AGBW02014485 OWR41592.1 ODYU01006144 SOQ47743.1 NWSH01003396 PCG66374.1 PCG66373.1 ODYU01011083 SOQ56599.1 ODYU01003605 SOQ42578.1 PZC78978.1 PZC78979.1 NWSH01005898 PCG63860.1 PZC78987.1 BR000604 FAA00606.1 ODYU01003855 SOQ43136.1 BR000603 FAA00605.1 BABH01013344 BR000605 FAA00607.1 AB047477 BAB32475.1 PZC78984.1 AB047478 BAB32476.1 AK402581 BAM19203.1 BR000602 FAA00604.1 BR000606 FAA00608.1 AK401621 BAM18243.1 AK402491 BAM19113.1 BABH01013339 BR000588 FAA00590.1 BR000587 FAA00589.1 AGBW02009770 OWR49962.1 AK401616 BAM18238.1 KPI97790.1 KPI97791.1 KPJ09758.1 KPJ09766.1 KPI97781.1 OWR49961.1 AGBW02009537 OWR50562.1 KPJ09759.1
FAA00600.1 BR000596 FAA00598.1 AB047481 BAB32478.1 BR000600 FAA00602.1 BR000599 FAA00601.1 AB047480 BAB32477.1 AB264676 BAG30752.1 KQ459586 KPI97786.1 AK405036 BAM20443.1 AK401509 BAM18131.1 AGBW02010913 OWR47527.1 KQ461073 KPJ09764.1 KPJ09765.1 KZ149892 PZC78981.1 NWSH01003366 PCG66457.1 AK405035 BAM20442.1 OWR47528.1 PZC78982.1 PZC78985.1 AK401842 BAM18464.1 KPI97785.1 AB264683 BAG30759.1 PZC78988.1 KPI97784.1 PZC78983.1 KQ460333 KPJ15643.1 AGBW02014485 OWR41592.1 ODYU01006144 SOQ47743.1 NWSH01003396 PCG66374.1 PCG66373.1 ODYU01011083 SOQ56599.1 ODYU01003605 SOQ42578.1 PZC78978.1 PZC78979.1 NWSH01005898 PCG63860.1 PZC78987.1 BR000604 FAA00606.1 ODYU01003855 SOQ43136.1 BR000603 FAA00605.1 BABH01013344 BR000605 FAA00607.1 AB047477 BAB32475.1 PZC78984.1 AB047478 BAB32476.1 AK402581 BAM19203.1 BR000602 FAA00604.1 BR000606 FAA00608.1 AK401621 BAM18243.1 AK402491 BAM19113.1 BABH01013339 BR000588 FAA00590.1 BR000587 FAA00589.1 AGBW02009770 OWR49962.1 AK401616 BAM18238.1 KPI97790.1 KPI97791.1 KPJ09758.1 KPJ09766.1 KPI97781.1 OWR49961.1 AGBW02009537 OWR50562.1 KPJ09759.1
Proteomes
Pfam
PF00379 Chitin_bind_4
ProteinModelPortal
C0H6U2
H9ISN7
C0H6T8
C0H6T9
C0H6T7
Q9BPR6
+ More
C0H6U1 C0H6U0 Q9BPR7 B2DBJ3 A0A194PWP3 I4DRA3 I4DJP1 A0A212F1C1 A0A194QWT1 A0A194QXS8 A0A2W1BXD1 A0A2A4J2S9 I4DRA2 A0A212F1A8 A0A2W1BV71 A0A2W1BZV6 I4DKM4 A0A194Q302 B2DBK0 A0A2W1BVB0 A0A194PYF6 A0A2W1C485 A0A0N1IPH5 A0A212EJB4 A0A2H1W3Y2 A0A2A4J4D6 A0A2A4J4N1 A0A2H1WU56 A0A2H1VP46 A0A2W1BVH0 A0A2A4IXI8 A0A2W1C0G8 C0H6U5 A0A2H1VQS4 H9IT50 C0H6U4 C0H6U6 Q9BPR9 A0A2W1BZH0 Q9BPR8 I4DMR3 C0H6U3 C0H6U7 I4DK03 I4DMH3 C0H6S9 C0H6S8 A0A212F891 I4DJZ8 A0A194PWQ0 A0A194QW46 A0A194R1N5 A0A194PWP0 A0A212F8A9 A0A212F9Z8 A0A194QWS6
C0H6U1 C0H6U0 Q9BPR7 B2DBJ3 A0A194PWP3 I4DRA3 I4DJP1 A0A212F1C1 A0A194QWT1 A0A194QXS8 A0A2W1BXD1 A0A2A4J2S9 I4DRA2 A0A212F1A8 A0A2W1BV71 A0A2W1BZV6 I4DKM4 A0A194Q302 B2DBK0 A0A2W1BVB0 A0A194PYF6 A0A2W1C485 A0A0N1IPH5 A0A212EJB4 A0A2H1W3Y2 A0A2A4J4D6 A0A2A4J4N1 A0A2H1WU56 A0A2H1VP46 A0A2W1BVH0 A0A2A4IXI8 A0A2W1C0G8 C0H6U5 A0A2H1VQS4 H9IT50 C0H6U4 C0H6U6 Q9BPR9 A0A2W1BZH0 Q9BPR8 I4DMR3 C0H6U3 C0H6U7 I4DK03 I4DMH3 C0H6S9 C0H6S8 A0A212F891 I4DJZ8 A0A194PWQ0 A0A194QW46 A0A194R1N5 A0A194PWP0 A0A212F8A9 A0A212F9Z8 A0A194QWS6
Ontologies
GO
Topology
SignalP
Position: 1 - 21,
Likelihood: 0.960719
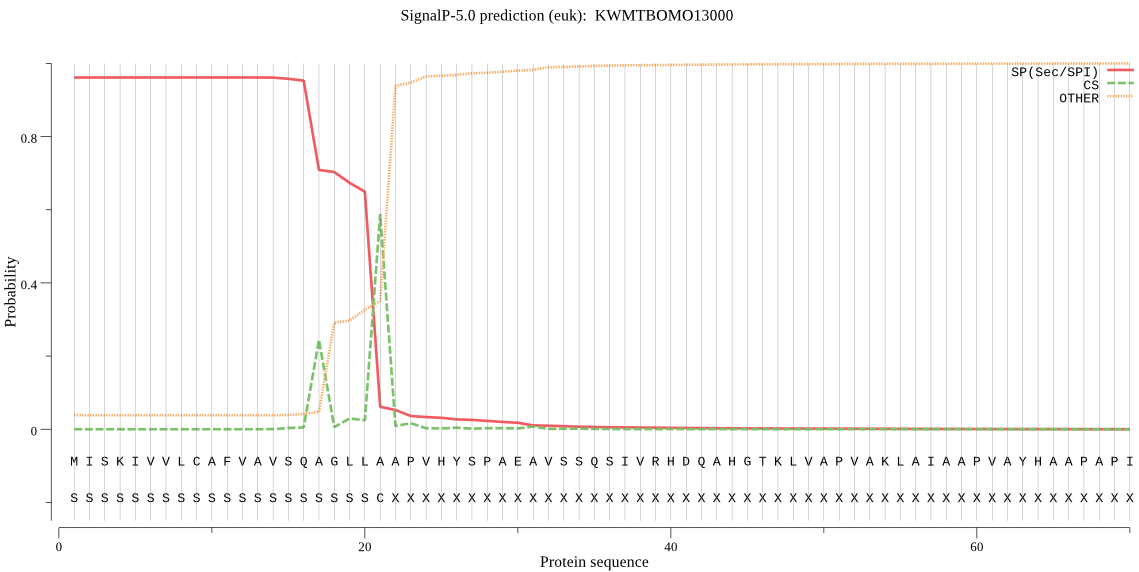
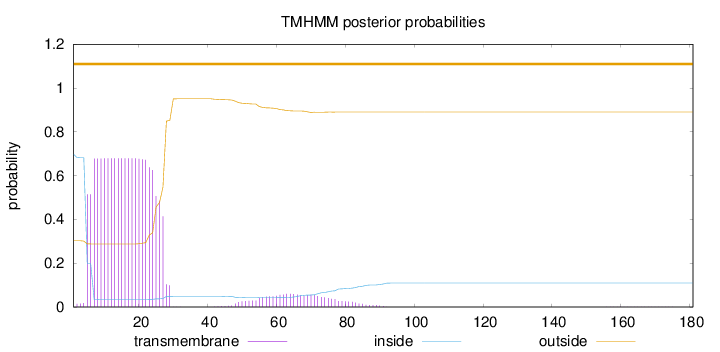
Length:
181
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
16.37559
Exp number, first 60 AAs:
15.27554
Total prob of N-in:
0.69753
POSSIBLE N-term signal
sequence
outside
1 - 181
Population Genetic Test Statistics
Pi
396.318343
Theta
244.339372
Tajima's D
1.945383
CLR
0.000922
CSRT
0.874056297185141
Interpretation
Uncertain
