Gene
KWMTBOMO12992
Pre Gene Modal
BGIBMGA000260
Annotation
cuticular_protein_RR-2_motif_107_precursor_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.457 Mitochondrial Reliability : 1.022 Nuclear Reliability : 1.403
Sequence
CDS
ATGTATTCCAAGGCTCATCCTAAATACGCCTTTGAGTACAAAGTAGAGGATCCTCACACCGGCGACAACAAGTACCAGCACGAGACCCGTGACGGTGACGTCGTGAAGGGCGTGTACAGTCTGCACGAGGCTGATGGTACCATCAGGACTGTTGAGTACAGCGCCGATAAACACAGCGGATTCAACGCAGTTGTGAGACGCGAGGGTCATGCTCACCATGCTGTACCTGAACATCATCATCATCACTAA
Protein
MYSKAHPKYAFEYKVEDPHTGDNKYQHETRDGDVVKGVYSLHEADGTIRTVEYSADKHSGFNAVVRREGHAHHAVPEHHHHH
Summary
Uniprot
C0H6U9
C0H6V1
C0H6V3
H9IT54
A0A3S2LRS0
A0A3S2TQL4
+ More
A0A3S2PIL6 C0H6W6 Q9BH30 A0A212FKA8 H9IT58 Q9BPR2 C0H6W4 A0A2H1WAB8 A0A2H1VRQ2 A0A2H1VPL7 I4DID6 A0A194PWM9 A0A194QWC3 C0H6W5 A0A2H1VFG1 H9IT59 A0A2W1BXC1 A0A2H1VFK7 A0A2W1C1G9 A0A2H1WWL0 A0A067RMC7 A0A2H1VH24 A0A2H1VQ88 A0A2H1WE29 A0A2H1WJC4 A0A0L7KHQ9 A0A2H1V127 D6WJ00 A0A194QWU2 A0A194PXM4 A0A194PYE7 A0A1Y1KZI1 D6WJ01 A0A2H1V4C4 A0A194QXT8 A0A2P8Y145 A0A2W1BV53 A0A2H1VQS4 A0A194R1P3 A0A2P8Y151 A0A2H1VQR4 A0A3S2NYR7 D6WIZ9 I4DKM3 C0H6W8 A0A2R7VRX6 A0A2H1VPL0 Q16R88 B2DBJ3 A0A1J1IKJ9 A0A2H1WU56 G8FVP8 A0A2W1BZF1 A0A2H1WF17 A0A2H1VPE0 A0A194PWP3 A0A2W1BXB1 I4DJP1 I4DRA3 A0A2H1W9R4 A0A2W1BZU7 A0A069DNE3 R4G2R9 A0A2A3ESS6 A0A194QWT1 T1HRQ0 A0A1S3DBS1 A0A194Q302 U4TRW7 A0A2A4J906 A0A2H1VPH0 A0A182TZH3 A0A182X2B9 A7URY7 A0A182I6I1 A0A182N9I2 A0A2S1ZS96 A0A182Q5D4 A0A182MBA2 A0A182WBA9 A0A2P8Y134 A0A2S1ZS95 W5JT23 R4FNZ4 T1I2B8 A0A2H1WDQ1 I4DKM1 A0A2H1WXM0 T1DSN0 A0A194PYE3
A0A3S2PIL6 C0H6W6 Q9BH30 A0A212FKA8 H9IT58 Q9BPR2 C0H6W4 A0A2H1WAB8 A0A2H1VRQ2 A0A2H1VPL7 I4DID6 A0A194PWM9 A0A194QWC3 C0H6W5 A0A2H1VFG1 H9IT59 A0A2W1BXC1 A0A2H1VFK7 A0A2W1C1G9 A0A2H1WWL0 A0A067RMC7 A0A2H1VH24 A0A2H1VQ88 A0A2H1WE29 A0A2H1WJC4 A0A0L7KHQ9 A0A2H1V127 D6WJ00 A0A194QWU2 A0A194PXM4 A0A194PYE7 A0A1Y1KZI1 D6WJ01 A0A2H1V4C4 A0A194QXT8 A0A2P8Y145 A0A2W1BV53 A0A2H1VQS4 A0A194R1P3 A0A2P8Y151 A0A2H1VQR4 A0A3S2NYR7 D6WIZ9 I4DKM3 C0H6W8 A0A2R7VRX6 A0A2H1VPL0 Q16R88 B2DBJ3 A0A1J1IKJ9 A0A2H1WU56 G8FVP8 A0A2W1BZF1 A0A2H1WF17 A0A2H1VPE0 A0A194PWP3 A0A2W1BXB1 I4DJP1 I4DRA3 A0A2H1W9R4 A0A2W1BZU7 A0A069DNE3 R4G2R9 A0A2A3ESS6 A0A194QWT1 T1HRQ0 A0A1S3DBS1 A0A194Q302 U4TRW7 A0A2A4J906 A0A2H1VPH0 A0A182TZH3 A0A182X2B9 A7URY7 A0A182I6I1 A0A182N9I2 A0A2S1ZS96 A0A182Q5D4 A0A182MBA2 A0A182WBA9 A0A2P8Y134 A0A2S1ZS95 W5JT23 R4FNZ4 T1I2B8 A0A2H1WDQ1 I4DKM1 A0A2H1WXM0 T1DSN0 A0A194PYE3
Pubmed
EMBL
BABH01013348
BR000608
FAA00610.1
BR000610
FAA00612.1
BR000612
+ More
FAA00614.1 RSAL01000021 RVE52446.1 RVE52443.1 RVE52447.1 BR000625 FAA00627.1 AB047485 AB047486 BAB32482.1 BAB32483.1 AGBW02008093 OWR54166.1 BABH01013354 AB047487 BAB32484.1 BR000623 FAA00625.1 ODYU01007211 SOQ49792.1 ODYU01004033 SOQ43499.1 ODYU01003691 SOQ42770.1 AK401054 BAM17676.1 KQ459586 KPI97771.1 KQ461073 KPJ09777.1 BR000624 FAA00626.1 ODYU01002284 SOQ39565.1 BABH01013355 KZ149892 PZC78971.1 SOQ39566.1 PZC78970.1 ODYU01011607 SOQ57450.1 KK852584 KDR20834.1 SOQ39564.1 ODYU01003629 SOQ42622.1 ODYU01007979 SOQ51206.1 ODYU01009022 SOQ53117.1 JTDY01009813 KOB62611.1 ODYU01000177 SOQ34521.1 KQ971342 EFA03719.1 KPJ09774.1 KPI97773.1 KPI97774.1 GEZM01068802 JAV66764.1 EFA03718.1 ODYU01000605 SOQ35681.1 KPJ09775.1 PYGN01001062 PSN37960.1 PZC78962.1 ODYU01003855 SOQ43136.1 KPJ09776.1 PSN37969.1 SOQ43137.1 RVE52458.1 EFA03720.1 AK401841 BAM18463.1 BABH01013358 BR000627 FAA00629.1 KK854044 PTY10186.1 SOQ42769.1 CH477721 EAT36915.1 AB264676 BAG30752.1 CVRI01000054 CRL00060.1 ODYU01011083 SOQ56599.1 JN082718 AER27815.1 PZC78964.1 ODYU01008253 SOQ51680.1 ODYU01003654 SOQ42695.1 KPI97786.1 PZC78961.1 AK401509 BAM18131.1 AK405036 BAM20443.1 SOQ49793.1 PZC78975.1 GBGD01003554 JAC85335.1 ACPB03012170 GAHY01002361 JAA75149.1 KZ288186 PBC34778.1 KPJ09764.1 ACPB03012169 KPI97785.1 KB631296 ERL84224.1 NWSH01002544 PCG68014.1 SOQ42696.1 AAAB01008807 EDO64780.1 APCN01003642 MF942810 AWK28333.1 AXCN02000565 AXCM01020208 PSN37956.1 MF942812 AWK28335.1 ADMH02000233 ETN67291.1 GAHY01000373 JAA77137.1 ACPB03022691 SOQ51205.1 AK401839 BAM18461.1 ODYU01011838 SOQ57809.1 GAMD01001404 JAB00187.1 KPI97769.1
FAA00614.1 RSAL01000021 RVE52446.1 RVE52443.1 RVE52447.1 BR000625 FAA00627.1 AB047485 AB047486 BAB32482.1 BAB32483.1 AGBW02008093 OWR54166.1 BABH01013354 AB047487 BAB32484.1 BR000623 FAA00625.1 ODYU01007211 SOQ49792.1 ODYU01004033 SOQ43499.1 ODYU01003691 SOQ42770.1 AK401054 BAM17676.1 KQ459586 KPI97771.1 KQ461073 KPJ09777.1 BR000624 FAA00626.1 ODYU01002284 SOQ39565.1 BABH01013355 KZ149892 PZC78971.1 SOQ39566.1 PZC78970.1 ODYU01011607 SOQ57450.1 KK852584 KDR20834.1 SOQ39564.1 ODYU01003629 SOQ42622.1 ODYU01007979 SOQ51206.1 ODYU01009022 SOQ53117.1 JTDY01009813 KOB62611.1 ODYU01000177 SOQ34521.1 KQ971342 EFA03719.1 KPJ09774.1 KPI97773.1 KPI97774.1 GEZM01068802 JAV66764.1 EFA03718.1 ODYU01000605 SOQ35681.1 KPJ09775.1 PYGN01001062 PSN37960.1 PZC78962.1 ODYU01003855 SOQ43136.1 KPJ09776.1 PSN37969.1 SOQ43137.1 RVE52458.1 EFA03720.1 AK401841 BAM18463.1 BABH01013358 BR000627 FAA00629.1 KK854044 PTY10186.1 SOQ42769.1 CH477721 EAT36915.1 AB264676 BAG30752.1 CVRI01000054 CRL00060.1 ODYU01011083 SOQ56599.1 JN082718 AER27815.1 PZC78964.1 ODYU01008253 SOQ51680.1 ODYU01003654 SOQ42695.1 KPI97786.1 PZC78961.1 AK401509 BAM18131.1 AK405036 BAM20443.1 SOQ49793.1 PZC78975.1 GBGD01003554 JAC85335.1 ACPB03012170 GAHY01002361 JAA75149.1 KZ288186 PBC34778.1 KPJ09764.1 ACPB03012169 KPI97785.1 KB631296 ERL84224.1 NWSH01002544 PCG68014.1 SOQ42696.1 AAAB01008807 EDO64780.1 APCN01003642 MF942810 AWK28333.1 AXCN02000565 AXCM01020208 PSN37956.1 MF942812 AWK28335.1 ADMH02000233 ETN67291.1 GAHY01000373 JAA77137.1 ACPB03022691 SOQ51205.1 AK401839 BAM18461.1 ODYU01011838 SOQ57809.1 GAMD01001404 JAB00187.1 KPI97769.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053268
UP000053240
UP000027135
+ More
UP000037510 UP000007266 UP000245037 UP000008820 UP000183832 UP000015103 UP000242457 UP000079169 UP000030742 UP000218220 UP000075902 UP000076407 UP000007062 UP000075840 UP000075884 UP000075886 UP000075883 UP000075920 UP000000673
UP000037510 UP000007266 UP000245037 UP000008820 UP000183832 UP000015103 UP000242457 UP000079169 UP000030742 UP000218220 UP000075902 UP000076407 UP000007062 UP000075840 UP000075884 UP000075886 UP000075883 UP000075920 UP000000673
Interpro
Gene 3D
ProteinModelPortal
C0H6U9
C0H6V1
C0H6V3
H9IT54
A0A3S2LRS0
A0A3S2TQL4
+ More
A0A3S2PIL6 C0H6W6 Q9BH30 A0A212FKA8 H9IT58 Q9BPR2 C0H6W4 A0A2H1WAB8 A0A2H1VRQ2 A0A2H1VPL7 I4DID6 A0A194PWM9 A0A194QWC3 C0H6W5 A0A2H1VFG1 H9IT59 A0A2W1BXC1 A0A2H1VFK7 A0A2W1C1G9 A0A2H1WWL0 A0A067RMC7 A0A2H1VH24 A0A2H1VQ88 A0A2H1WE29 A0A2H1WJC4 A0A0L7KHQ9 A0A2H1V127 D6WJ00 A0A194QWU2 A0A194PXM4 A0A194PYE7 A0A1Y1KZI1 D6WJ01 A0A2H1V4C4 A0A194QXT8 A0A2P8Y145 A0A2W1BV53 A0A2H1VQS4 A0A194R1P3 A0A2P8Y151 A0A2H1VQR4 A0A3S2NYR7 D6WIZ9 I4DKM3 C0H6W8 A0A2R7VRX6 A0A2H1VPL0 Q16R88 B2DBJ3 A0A1J1IKJ9 A0A2H1WU56 G8FVP8 A0A2W1BZF1 A0A2H1WF17 A0A2H1VPE0 A0A194PWP3 A0A2W1BXB1 I4DJP1 I4DRA3 A0A2H1W9R4 A0A2W1BZU7 A0A069DNE3 R4G2R9 A0A2A3ESS6 A0A194QWT1 T1HRQ0 A0A1S3DBS1 A0A194Q302 U4TRW7 A0A2A4J906 A0A2H1VPH0 A0A182TZH3 A0A182X2B9 A7URY7 A0A182I6I1 A0A182N9I2 A0A2S1ZS96 A0A182Q5D4 A0A182MBA2 A0A182WBA9 A0A2P8Y134 A0A2S1ZS95 W5JT23 R4FNZ4 T1I2B8 A0A2H1WDQ1 I4DKM1 A0A2H1WXM0 T1DSN0 A0A194PYE3
A0A3S2PIL6 C0H6W6 Q9BH30 A0A212FKA8 H9IT58 Q9BPR2 C0H6W4 A0A2H1WAB8 A0A2H1VRQ2 A0A2H1VPL7 I4DID6 A0A194PWM9 A0A194QWC3 C0H6W5 A0A2H1VFG1 H9IT59 A0A2W1BXC1 A0A2H1VFK7 A0A2W1C1G9 A0A2H1WWL0 A0A067RMC7 A0A2H1VH24 A0A2H1VQ88 A0A2H1WE29 A0A2H1WJC4 A0A0L7KHQ9 A0A2H1V127 D6WJ00 A0A194QWU2 A0A194PXM4 A0A194PYE7 A0A1Y1KZI1 D6WJ01 A0A2H1V4C4 A0A194QXT8 A0A2P8Y145 A0A2W1BV53 A0A2H1VQS4 A0A194R1P3 A0A2P8Y151 A0A2H1VQR4 A0A3S2NYR7 D6WIZ9 I4DKM3 C0H6W8 A0A2R7VRX6 A0A2H1VPL0 Q16R88 B2DBJ3 A0A1J1IKJ9 A0A2H1WU56 G8FVP8 A0A2W1BZF1 A0A2H1WF17 A0A2H1VPE0 A0A194PWP3 A0A2W1BXB1 I4DJP1 I4DRA3 A0A2H1W9R4 A0A2W1BZU7 A0A069DNE3 R4G2R9 A0A2A3ESS6 A0A194QWT1 T1HRQ0 A0A1S3DBS1 A0A194Q302 U4TRW7 A0A2A4J906 A0A2H1VPH0 A0A182TZH3 A0A182X2B9 A7URY7 A0A182I6I1 A0A182N9I2 A0A2S1ZS96 A0A182Q5D4 A0A182MBA2 A0A182WBA9 A0A2P8Y134 A0A2S1ZS95 W5JT23 R4FNZ4 T1I2B8 A0A2H1WDQ1 I4DKM1 A0A2H1WXM0 T1DSN0 A0A194PYE3
Ontologies
GO
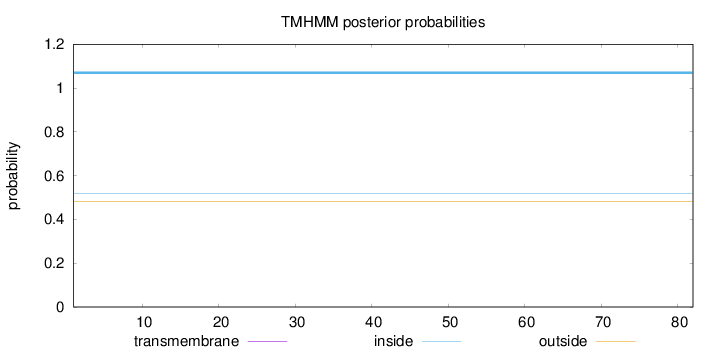
Topology
Length:
82
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.51850
inside
1 - 82
Population Genetic Test Statistics
Pi
406.825549
Theta
202.08869
Tajima's D
3.264851
CLR
0.349366
CSRT
0.988900554972251
Interpretation
Uncertain
