Gene
KWMTBOMO12985
Pre Gene Modal
BGIBMGA000253
Annotation
cuticular_protein_RR-2_motif_121_precursor_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 2.225
Sequence
CDS
ATGTTCTCAAAGATTGTATGTCTACTTGGTGTTGTTGCTGTTGCAACTGCGCAGTATGGTCATGACCAAAGTCACGGCCACGCCTTCTCGTCGCAACACATCTCGCGGCACGATGGCCCCGCACAAGTTGTGCATGTCAAGGGTCACGGGCACGATGGTCATGGTCACGAAGACCACATCGACTACTATGCCCACCCTAAATACGAGTTCGAGTACAAAGTGAGCGACCCTCACACCGGCGACCACAAGAGTCAGCACGAGTCCCGCGACGGTGACGTCGTCAAGGGATATTACTCGCTGCATCAACCTGACGGTTCCATCAGGCATGTCGACTATCACGGCGACAAGCACTCAGGATTCCATGCCACCGTGAAGCACAGCACCCACCACATTGCCCCTGAAAAGCACCATCAGTAA
Protein
MFSKIVCLLGVVAVATAQYGHDQSHGHAFSSQHISRHDGPAQVVHVKGHGHDGHGHEDHIDYYAHPKYEFEYKVSDPHTGDHKSQHESRDGDVVKGYYSLHQPDGSIRHVDYHGDKHSGFHATVKHSTHHIAPEKHHQ
Summary
Uniprot
C0H6W3
A0A194PXM4
A0A2H1VP60
A0A194R1P3
A0A194R1N8
A0A2W1C0C0
+ More
A0A2A4IV03 G8FVQ2 A0A194PWN4 C0H6W0 A0A194QW60 C0H6V6 C0H6V5 C0H6V7 C0H6V4 A0A2H1WGD3 A0A194PXM8 A0A2H1WRJ6 A0A2W1BVG0 C0H6V8 C0H6U8 A0A2H1WDQ1 A0A3S2PIL6 A0A2H1VQR4 A0A2W1C1X5 C0H6W1 C0H6V0 A0A3S2LRS0 A0A2H1VPL7 C0H6V2 A0A194PWS5 A0A2H1V127 Q9BH30 H9ISM7 A0A2W1C1W6 A0A2W1BXC1 A0A194QWB8 A0A2H1VQS4 A0A212F8A2 A0A2W1BZW6 I4DMR3 D6WJ08 A0A194PXP1 A0A194QWA3 U4U2J7 A0A3S2NJA9 I4DMH3 A0A2W1BZU2 A0A1W4WQB0 C0H6S9 C0H6S8 A0A194PWU5 A0A194PWQ0 A0A2H1VD38 A0A212F899 A0A194PWT0 A0A212F897 A0A194PYF2 I4DJZ8 N6TM36 A0A182N9I2
A0A2A4IV03 G8FVQ2 A0A194PWN4 C0H6W0 A0A194QW60 C0H6V6 C0H6V5 C0H6V7 C0H6V4 A0A2H1WGD3 A0A194PXM8 A0A2H1WRJ6 A0A2W1BVG0 C0H6V8 C0H6U8 A0A2H1WDQ1 A0A3S2PIL6 A0A2H1VQR4 A0A2W1C1X5 C0H6W1 C0H6V0 A0A3S2LRS0 A0A2H1VPL7 C0H6V2 A0A194PWS5 A0A2H1V127 Q9BH30 H9ISM7 A0A2W1C1W6 A0A2W1BXC1 A0A194QWB8 A0A2H1VQS4 A0A212F8A2 A0A2W1BZW6 I4DMR3 D6WJ08 A0A194PXP1 A0A194QWA3 U4U2J7 A0A3S2NJA9 I4DMH3 A0A2W1BZU2 A0A1W4WQB0 C0H6S9 C0H6S8 A0A194PWU5 A0A194PWQ0 A0A2H1VD38 A0A212F899 A0A194PWT0 A0A212F897 A0A194PYF2 I4DJZ8 N6TM36 A0A182N9I2
Pubmed
EMBL
BABH01013354
BR000622
FAA00624.1
KQ459586
KPI97773.1
ODYU01003629
+ More
SOQ42621.1 KQ461073 KPJ09776.1 KPJ09771.1 KZ149892 PZC78967.1 NWSH01006625 PCG63326.1 JN082722 AER27819.1 KPI97776.1 BR000619 FAA00621.1 KPJ09773.1 BABH01013350 BR000615 FAA00617.1 BABH01013349 BR000614 FAA00616.1 BABH01013351 BR000616 BR000618 FAA00618.1 FAA00620.1 BR000613 FAA00615.1 ODYU01008423 SOQ52006.1 KPI97778.1 ODYU01010466 SOQ55596.1 PZC78969.1 BR000617 FAA00619.1 BABH01013347 BR000607 FAA00609.1 ODYU01007979 SOQ51205.1 RSAL01000021 RVE52447.1 ODYU01003855 SOQ43137.1 PZC78976.1 BR000620 FAA00622.1 BR000609 FAA00611.1 RVE52446.1 ODYU01003691 SOQ42770.1 BR000611 FAA00613.1 KPI97772.1 ODYU01000177 SOQ34521.1 AB047485 AB047486 BAB32482.1 BAB32483.1 PZC78966.1 PZC78971.1 KPJ09772.1 SOQ43136.1 AGBW02009770 OWR49967.1 PZC78995.1 AK402581 BAM19203.1 KQ971342 EFA03712.1 KPI97793.1 KPJ09757.1 KB631296 ERL84225.1 RVE52429.1 AK402491 BAM19113.1 PZC78965.1 BABH01013339 BR000588 FAA00590.1 BR000587 FAA00589.1 KPI97792.1 KPI97791.1 ODYU01001904 SOQ38763.1 OWR49965.1 KPI97777.1 OWR49966.1 KPI97779.1 AK401616 BAM18238.1 KPI97790.1 APGK01018857 KB740085 KB631844 ENN81509.1 ERL84223.1 ERL86682.1
SOQ42621.1 KQ461073 KPJ09776.1 KPJ09771.1 KZ149892 PZC78967.1 NWSH01006625 PCG63326.1 JN082722 AER27819.1 KPI97776.1 BR000619 FAA00621.1 KPJ09773.1 BABH01013350 BR000615 FAA00617.1 BABH01013349 BR000614 FAA00616.1 BABH01013351 BR000616 BR000618 FAA00618.1 FAA00620.1 BR000613 FAA00615.1 ODYU01008423 SOQ52006.1 KPI97778.1 ODYU01010466 SOQ55596.1 PZC78969.1 BR000617 FAA00619.1 BABH01013347 BR000607 FAA00609.1 ODYU01007979 SOQ51205.1 RSAL01000021 RVE52447.1 ODYU01003855 SOQ43137.1 PZC78976.1 BR000620 FAA00622.1 BR000609 FAA00611.1 RVE52446.1 ODYU01003691 SOQ42770.1 BR000611 FAA00613.1 KPI97772.1 ODYU01000177 SOQ34521.1 AB047485 AB047486 BAB32482.1 BAB32483.1 PZC78966.1 PZC78971.1 KPJ09772.1 SOQ43136.1 AGBW02009770 OWR49967.1 PZC78995.1 AK402581 BAM19203.1 KQ971342 EFA03712.1 KPI97793.1 KPJ09757.1 KB631296 ERL84225.1 RVE52429.1 AK402491 BAM19113.1 PZC78965.1 BABH01013339 BR000588 FAA00590.1 BR000587 FAA00589.1 KPI97792.1 KPI97791.1 ODYU01001904 SOQ38763.1 OWR49965.1 KPI97777.1 OWR49966.1 KPI97779.1 AK401616 BAM18238.1 KPI97790.1 APGK01018857 KB740085 KB631844 ENN81509.1 ERL84223.1 ERL86682.1
Proteomes
Pfam
PF00379 Chitin_bind_4
ProteinModelPortal
C0H6W3
A0A194PXM4
A0A2H1VP60
A0A194R1P3
A0A194R1N8
A0A2W1C0C0
+ More
A0A2A4IV03 G8FVQ2 A0A194PWN4 C0H6W0 A0A194QW60 C0H6V6 C0H6V5 C0H6V7 C0H6V4 A0A2H1WGD3 A0A194PXM8 A0A2H1WRJ6 A0A2W1BVG0 C0H6V8 C0H6U8 A0A2H1WDQ1 A0A3S2PIL6 A0A2H1VQR4 A0A2W1C1X5 C0H6W1 C0H6V0 A0A3S2LRS0 A0A2H1VPL7 C0H6V2 A0A194PWS5 A0A2H1V127 Q9BH30 H9ISM7 A0A2W1C1W6 A0A2W1BXC1 A0A194QWB8 A0A2H1VQS4 A0A212F8A2 A0A2W1BZW6 I4DMR3 D6WJ08 A0A194PXP1 A0A194QWA3 U4U2J7 A0A3S2NJA9 I4DMH3 A0A2W1BZU2 A0A1W4WQB0 C0H6S9 C0H6S8 A0A194PWU5 A0A194PWQ0 A0A2H1VD38 A0A212F899 A0A194PWT0 A0A212F897 A0A194PYF2 I4DJZ8 N6TM36 A0A182N9I2
A0A2A4IV03 G8FVQ2 A0A194PWN4 C0H6W0 A0A194QW60 C0H6V6 C0H6V5 C0H6V7 C0H6V4 A0A2H1WGD3 A0A194PXM8 A0A2H1WRJ6 A0A2W1BVG0 C0H6V8 C0H6U8 A0A2H1WDQ1 A0A3S2PIL6 A0A2H1VQR4 A0A2W1C1X5 C0H6W1 C0H6V0 A0A3S2LRS0 A0A2H1VPL7 C0H6V2 A0A194PWS5 A0A2H1V127 Q9BH30 H9ISM7 A0A2W1C1W6 A0A2W1BXC1 A0A194QWB8 A0A2H1VQS4 A0A212F8A2 A0A2W1BZW6 I4DMR3 D6WJ08 A0A194PXP1 A0A194QWA3 U4U2J7 A0A3S2NJA9 I4DMH3 A0A2W1BZU2 A0A1W4WQB0 C0H6S9 C0H6S8 A0A194PWU5 A0A194PWQ0 A0A2H1VD38 A0A212F899 A0A194PWT0 A0A212F897 A0A194PYF2 I4DJZ8 N6TM36 A0A182N9I2
Ontologies
GO
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.986560
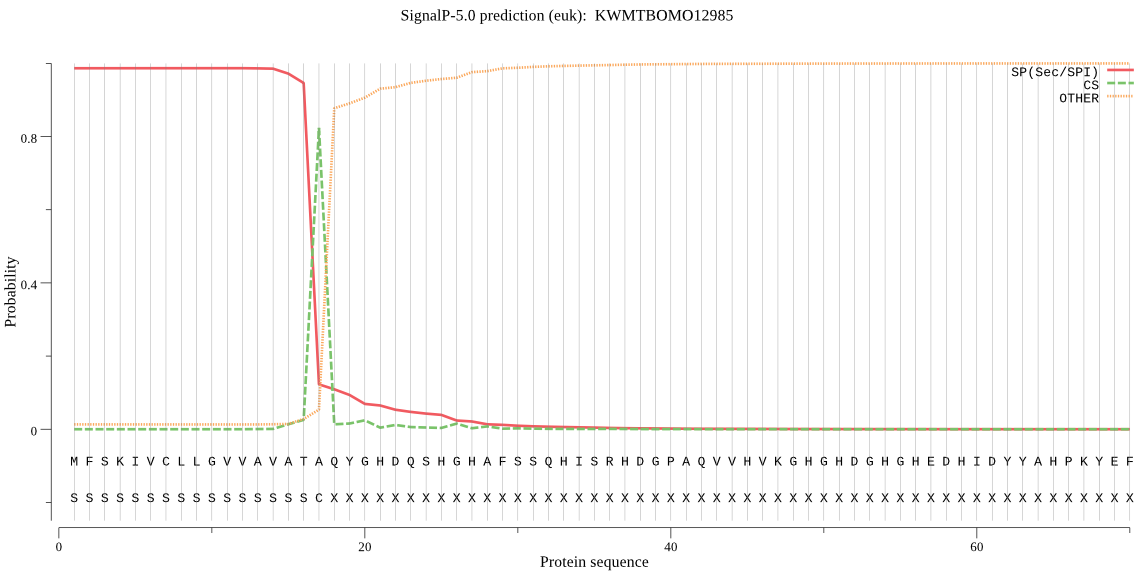
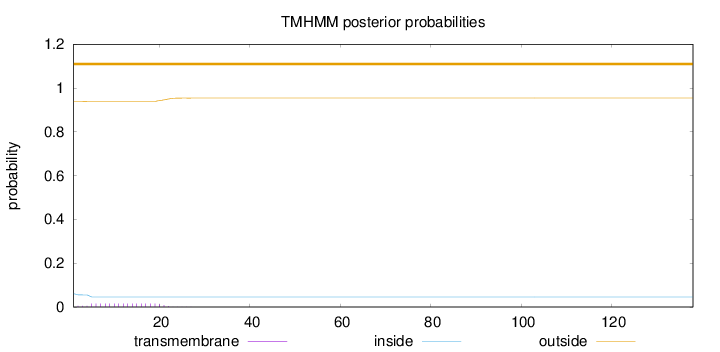
Length:
138
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.27628
Exp number, first 60 AAs:
0.27628
Total prob of N-in:
0.06089
outside
1 - 138
Population Genetic Test Statistics
Pi
21.889568
Theta
25.690124
Tajima's D
-0.502327
CLR
2.035052
CSRT
0.242937853107345
Interpretation
Uncertain
