Gene
KWMTBOMO12974
Pre Gene Modal
BGIBMGA000439
Annotation
PREDICTED:_band_7_protein_AGAP004871-like_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.671
Sequence
CDS
ATGGATTTCGGTTGCCACGACAAGGACTCAGGGGAAGTGGAGTGCTACTTCGAAATACATTGCGATACGGGTACTTCTTGCGCTATCACGATACAAATGCCGAACCCAGAAGCCGTTGGATGTGTAGAACGATTTGCAACTTTACTGTCATATCTACTTGTCATCATCACCTTCCCTTTTTCGTTATTTGAATGTTTTAAGGTTGTGCAAGAATTTGAAAGAGCTGTAATCTTCAGACTAGGCAGATTAAGGAAAGGCGGCGCGAGAGGCCCAGGACTATTCTTTGTTCTTCCCTGCATAGACACTTATAGAAAAGTAGATCTACGAACGGTTTCGTTTGACGTACCACCACAAGAGGTACTAACGCGAGATTCTGTGACAGTGGCAGTGGACGCTGTAGTGTATTACCGAATCAAAGAACCCTTAAATGCAGTGGTGAGAGTTGCAGACTATAGCGCGTCAACAAGACTCTTGGCCGCTACAACATTAAGGAATGTTCTTGGAATGAGGGATCTAGCGCAACTCCTCTCTGACAGGGAAGCAATCAGTCACATGATGCAAGCTAGCCTGGATGAGGCCACGGAGCCTTGGGGTGTCGAAGTAGAAAGAGTTGAAATTAAAGACGTCCGACTGCCCGTGCAGCTACAGAAGGCCATGGCAGCAGAAGCTGAGGCGGATCGTGAAGCAAGAGCGAAAATCATTGCCGCCGAAGGAGAAATTAAAGCATCAAAAGCATTGAAGGAAGCATCACTTGTCATGATCGACAATCCTATGGCCTTACAGCTGCGCTACCTGCAGTCTTTGAGCTCTATATCAGCAGAGAAGAATTCCACGATAATCTTTCCATTCCCTATGGATTTCTTGAAAGCATTTTTGGGAGGAGCCGGACCAAGTACAACGCACCAACCAAGCGTACAAATGACACAATCATTACCAATATAG
Protein
MDFGCHDKDSGEVECYFEIHCDTGTSCAITIQMPNPEAVGCVERFATLLSYLLVIITFPFSLFECFKVVQEFERAVIFRLGRLRKGGARGPGLFFVLPCIDTYRKVDLRTVSFDVPPQEVLTRDSVTVAVDAVVYYRIKEPLNAVVRVADYSASTRLLAATTLRNVLGMRDLAQLLSDREAISHMMQASLDEATEPWGVEVERVEIKDVRLPVQLQKAMAAEAEADREARAKIIAAEGEIKASKALKEASLVMIDNPMALQLRYLQSLSSISAEKNSTIIFPFPMDFLKAFLGGAGPSTTHQPSVQMTQSLPI
Summary
Uniprot
A0A2H1W1F3
A0A194QWD3
A0A3S2M712
H9IT60
A0A182J955
A0A182QUS2
+ More
A0A182LYX9 A0A226EN38 A0A088AUZ8 A0A1J1HFT6 A0A0P4W6L2 A0A182KBP2 V5GSQ7 A0A0P4W219 Q16P12 A0A023EPG9 A0A182XWM5 A0A2M4ANQ3 A0A182SLH3 Q7PCP9 A0A182HJL4 A0A182XKD0 A0A336K6L8 A0A182G0W2 A0A182P8N4 A0A2M4CMD3 F5HLR6 A0A1Q3G4I8 A0A1L8E079 A0A1L8E0D3 A0A0K8U3Q4 A0A1L8E0G5 A0A0A1X2W7 A0A182F7V9 W5JCU4 A0A0K8WIH9 A0A034WG10 A0A1B6BXA4 W8C0I7 A0A182NSR8 A0A067QRR3 F5HLR5 A0A182U4Y2 A0A182VY90 N6T1T2 A0A1B6J1H7 A0A1B6HNR5 J3JXA8 B4L4F0 K7IZ49 A0A084VUZ2 B4M2Q4 A0A0L0BQ17 A0A0A9WAT4 A0A1B6MBF9 A0A0P5C6B6 A0A1A9VV94 A0A1B0B115 A0A0A9WDU0 B4JJP0 A0A0L7QL39 A0A0P5D7T9 A0A0P5C3V9 E0VWU8 A0A1A9Z9R4 A0A0P5GG49 A0A224XI17 A0A0P5C684 A0A2R7WCH9 A0A1A9WVE6 A0A0P5C405 A0A0V0G688 B4NBZ3 A0A1B0FIT2 T1PAB0 A0A0P5CDF4 T1IAM2 A0A069DRQ9 A0A0P4VHG5 A0A161MZC9 A0A0P5CG43 A0A0N8AV13 A0A1I8Q9L0 A0A0P5C5K1 A0A023FCW0 A0A3S3PUL4 T1KPN5 A0A0P5C5K7 A0A0P5PL76 A0A0P4YJ31 A0A0P6EU95 A0A0P5EPL9 X2JFR0 A0A0K2UDU8 A0A0P5C5P6 A0A0P5CGY2 A0A0N8AKN4 B4I718 Q8MZ13
A0A182LYX9 A0A226EN38 A0A088AUZ8 A0A1J1HFT6 A0A0P4W6L2 A0A182KBP2 V5GSQ7 A0A0P4W219 Q16P12 A0A023EPG9 A0A182XWM5 A0A2M4ANQ3 A0A182SLH3 Q7PCP9 A0A182HJL4 A0A182XKD0 A0A336K6L8 A0A182G0W2 A0A182P8N4 A0A2M4CMD3 F5HLR6 A0A1Q3G4I8 A0A1L8E079 A0A1L8E0D3 A0A0K8U3Q4 A0A1L8E0G5 A0A0A1X2W7 A0A182F7V9 W5JCU4 A0A0K8WIH9 A0A034WG10 A0A1B6BXA4 W8C0I7 A0A182NSR8 A0A067QRR3 F5HLR5 A0A182U4Y2 A0A182VY90 N6T1T2 A0A1B6J1H7 A0A1B6HNR5 J3JXA8 B4L4F0 K7IZ49 A0A084VUZ2 B4M2Q4 A0A0L0BQ17 A0A0A9WAT4 A0A1B6MBF9 A0A0P5C6B6 A0A1A9VV94 A0A1B0B115 A0A0A9WDU0 B4JJP0 A0A0L7QL39 A0A0P5D7T9 A0A0P5C3V9 E0VWU8 A0A1A9Z9R4 A0A0P5GG49 A0A224XI17 A0A0P5C684 A0A2R7WCH9 A0A1A9WVE6 A0A0P5C405 A0A0V0G688 B4NBZ3 A0A1B0FIT2 T1PAB0 A0A0P5CDF4 T1IAM2 A0A069DRQ9 A0A0P4VHG5 A0A161MZC9 A0A0P5CG43 A0A0N8AV13 A0A1I8Q9L0 A0A0P5C5K1 A0A023FCW0 A0A3S3PUL4 T1KPN5 A0A0P5C5K7 A0A0P5PL76 A0A0P4YJ31 A0A0P6EU95 A0A0P5EPL9 X2JFR0 A0A0K2UDU8 A0A0P5C5P6 A0A0P5CGY2 A0A0N8AKN4 B4I718 Q8MZ13
Pubmed
26354079
19121390
17510324
24945155
25244985
12364791
+ More
14747013 17210077 26483478 25830018 20920257 23761445 25348373 24495485 24845553 23537049 22516182 17994087 20075255 24438588 26108605 25401762 20566863 25315136 26334808 27129103 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
14747013 17210077 26483478 25830018 20920257 23761445 25348373 24495485 24845553 23537049 22516182 17994087 20075255 24438588 26108605 25401762 20566863 25315136 26334808 27129103 25474469 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
ODYU01005723
SOQ46857.1
KQ461073
KPJ09787.1
RSAL01000021
RVE52466.1
+ More
BABH01013375 BABH01013376 AXCN02000227 AXCM01005935 LNIX01000003 OXA58444.1 CVRI01000001 CRK86418.1 GDRN01079140 JAI62455.1 GALX01003814 JAB64652.1 GDRN01079145 JAI62453.1 CH477800 EAT36089.1 GAPW01002697 JAC10901.1 GGFK01009084 MBW42405.1 AAAB01008904 EAA09668.5 UFQS01000071 UFQT01000071 SSW99005.1 SSX19387.1 JXUM01037796 KQ561142 KXJ79578.1 GGFL01002308 MBW66486.1 EGK97227.1 GFDL01000320 JAV34725.1 GFDF01001921 JAV12163.1 GFDF01001923 JAV12161.1 GDHF01031126 JAI21188.1 GFDF01001920 JAV12164.1 GBXI01009202 JAD05090.1 ADMH02001528 ETN62172.1 GDHF01001427 JAI50887.1 GAKP01004426 JAC54526.1 GEDC01031713 JAS05585.1 GAMC01001373 JAC05183.1 KK853400 KDR07856.1 EGK97226.1 APGK01055731 APGK01055732 KB741269 KB632330 ENN71473.1 ERL92552.1 GECU01034662 GECU01033952 GECU01017617 GECU01016518 GECU01014700 GECU01010920 GECU01007826 JAS73044.1 JAS73754.1 JAS90089.1 JAS91188.1 JAS93006.1 JAS96786.1 JAS99880.1 GECU01031380 JAS76326.1 BT127877 AEE62839.1 CH933810 EDW07428.2 ATLV01017130 ATLV01017131 ATLV01017132 ATLV01017133 ATLV01017134 KE525156 KFB41786.1 CH940651 EDW65958.2 JRES01001557 KNC22068.1 GBHO01038052 GBRD01003697 GBRD01003668 GBRD01003666 JAG05552.1 JAG62124.1 GEBQ01006748 JAT33229.1 GDIP01175590 JAJ47812.1 JXJN01006953 GBHO01038053 GBRD01011006 GBRD01011005 GBRD01003669 JAG05551.1 JAG54818.1 CH916370 EDV99792.1 KQ414934 KOC59294.1 GDIP01175941 JAJ47461.1 GDIP01175943 JAJ47459.1 DS235824 EEB17854.1 GDIQ01241408 JAK10317.1 GFTR01004299 JAW12127.1 GDIP01175636 JAJ47766.1 KK854455 PTY15995.1 GDIP01175944 JAJ47458.1 GECL01002540 JAP03584.1 CH964239 EDW82352.2 CCAG010016654 KA645629 AFP60258.1 GDIP01175940 JAJ47462.1 ACPB03005770 GBGD01002562 JAC86327.1 GDKW01002687 JAI53908.1 GEMB01003474 JAR99741.1 GDIP01175882 JAJ47520.1 GDIQ01240155 JAK11570.1 GDIP01175883 JAJ47519.1 GBBI01000203 JAC18509.1 NCKU01004031 NCKU01004020 NCKU01002970 NCKU01002969 RWS06541.1 RWS06561.1 RWS08438.1 RWS08439.1 CAEY01000333 GDIP01175637 JAJ47765.1 GDIQ01126895 JAL24831.1 GDIP01228035 JAI95366.1 GDIQ01059022 JAN35715.1 GDIQ01266386 JAJ85338.1 AE014298 AHN59898.1 HACA01018776 CDW36137.1 GDIP01175582 GDIP01175581 JAJ47820.1 GDIP01175580 JAJ47822.1 GDIQ01266387 JAJ85337.1 CH480823 EDW56116.1 AY113423 AAM29428.1 AAS65408.1
BABH01013375 BABH01013376 AXCN02000227 AXCM01005935 LNIX01000003 OXA58444.1 CVRI01000001 CRK86418.1 GDRN01079140 JAI62455.1 GALX01003814 JAB64652.1 GDRN01079145 JAI62453.1 CH477800 EAT36089.1 GAPW01002697 JAC10901.1 GGFK01009084 MBW42405.1 AAAB01008904 EAA09668.5 UFQS01000071 UFQT01000071 SSW99005.1 SSX19387.1 JXUM01037796 KQ561142 KXJ79578.1 GGFL01002308 MBW66486.1 EGK97227.1 GFDL01000320 JAV34725.1 GFDF01001921 JAV12163.1 GFDF01001923 JAV12161.1 GDHF01031126 JAI21188.1 GFDF01001920 JAV12164.1 GBXI01009202 JAD05090.1 ADMH02001528 ETN62172.1 GDHF01001427 JAI50887.1 GAKP01004426 JAC54526.1 GEDC01031713 JAS05585.1 GAMC01001373 JAC05183.1 KK853400 KDR07856.1 EGK97226.1 APGK01055731 APGK01055732 KB741269 KB632330 ENN71473.1 ERL92552.1 GECU01034662 GECU01033952 GECU01017617 GECU01016518 GECU01014700 GECU01010920 GECU01007826 JAS73044.1 JAS73754.1 JAS90089.1 JAS91188.1 JAS93006.1 JAS96786.1 JAS99880.1 GECU01031380 JAS76326.1 BT127877 AEE62839.1 CH933810 EDW07428.2 ATLV01017130 ATLV01017131 ATLV01017132 ATLV01017133 ATLV01017134 KE525156 KFB41786.1 CH940651 EDW65958.2 JRES01001557 KNC22068.1 GBHO01038052 GBRD01003697 GBRD01003668 GBRD01003666 JAG05552.1 JAG62124.1 GEBQ01006748 JAT33229.1 GDIP01175590 JAJ47812.1 JXJN01006953 GBHO01038053 GBRD01011006 GBRD01011005 GBRD01003669 JAG05551.1 JAG54818.1 CH916370 EDV99792.1 KQ414934 KOC59294.1 GDIP01175941 JAJ47461.1 GDIP01175943 JAJ47459.1 DS235824 EEB17854.1 GDIQ01241408 JAK10317.1 GFTR01004299 JAW12127.1 GDIP01175636 JAJ47766.1 KK854455 PTY15995.1 GDIP01175944 JAJ47458.1 GECL01002540 JAP03584.1 CH964239 EDW82352.2 CCAG010016654 KA645629 AFP60258.1 GDIP01175940 JAJ47462.1 ACPB03005770 GBGD01002562 JAC86327.1 GDKW01002687 JAI53908.1 GEMB01003474 JAR99741.1 GDIP01175882 JAJ47520.1 GDIQ01240155 JAK11570.1 GDIP01175883 JAJ47519.1 GBBI01000203 JAC18509.1 NCKU01004031 NCKU01004020 NCKU01002970 NCKU01002969 RWS06541.1 RWS06561.1 RWS08438.1 RWS08439.1 CAEY01000333 GDIP01175637 JAJ47765.1 GDIQ01126895 JAL24831.1 GDIP01228035 JAI95366.1 GDIQ01059022 JAN35715.1 GDIQ01266386 JAJ85338.1 AE014298 AHN59898.1 HACA01018776 CDW36137.1 GDIP01175582 GDIP01175581 JAJ47820.1 GDIP01175580 JAJ47822.1 GDIQ01266387 JAJ85337.1 CH480823 EDW56116.1 AY113423 AAM29428.1 AAS65408.1
Proteomes
UP000053240
UP000283053
UP000005204
UP000075880
UP000075886
UP000075883
+ More
UP000198287 UP000005203 UP000183832 UP000075881 UP000008820 UP000076408 UP000075901 UP000007062 UP000076407 UP000069940 UP000249989 UP000075885 UP000069272 UP000000673 UP000075884 UP000027135 UP000075902 UP000075920 UP000019118 UP000030742 UP000009192 UP000002358 UP000030765 UP000008792 UP000037069 UP000078200 UP000092460 UP000001070 UP000053825 UP000009046 UP000092445 UP000091820 UP000007798 UP000092444 UP000095301 UP000015103 UP000095300 UP000285301 UP000015104 UP000000803 UP000001292
UP000198287 UP000005203 UP000183832 UP000075881 UP000008820 UP000076408 UP000075901 UP000007062 UP000076407 UP000069940 UP000249989 UP000075885 UP000069272 UP000000673 UP000075884 UP000027135 UP000075902 UP000075920 UP000019118 UP000030742 UP000009192 UP000002358 UP000030765 UP000008792 UP000037069 UP000078200 UP000092460 UP000001070 UP000053825 UP000009046 UP000092445 UP000091820 UP000007798 UP000092444 UP000095301 UP000015103 UP000095300 UP000285301 UP000015104 UP000000803 UP000001292
Pfam
PF01145 Band_7
Interpro
SUPFAM
SSF117892
SSF117892
ProteinModelPortal
A0A2H1W1F3
A0A194QWD3
A0A3S2M712
H9IT60
A0A182J955
A0A182QUS2
+ More
A0A182LYX9 A0A226EN38 A0A088AUZ8 A0A1J1HFT6 A0A0P4W6L2 A0A182KBP2 V5GSQ7 A0A0P4W219 Q16P12 A0A023EPG9 A0A182XWM5 A0A2M4ANQ3 A0A182SLH3 Q7PCP9 A0A182HJL4 A0A182XKD0 A0A336K6L8 A0A182G0W2 A0A182P8N4 A0A2M4CMD3 F5HLR6 A0A1Q3G4I8 A0A1L8E079 A0A1L8E0D3 A0A0K8U3Q4 A0A1L8E0G5 A0A0A1X2W7 A0A182F7V9 W5JCU4 A0A0K8WIH9 A0A034WG10 A0A1B6BXA4 W8C0I7 A0A182NSR8 A0A067QRR3 F5HLR5 A0A182U4Y2 A0A182VY90 N6T1T2 A0A1B6J1H7 A0A1B6HNR5 J3JXA8 B4L4F0 K7IZ49 A0A084VUZ2 B4M2Q4 A0A0L0BQ17 A0A0A9WAT4 A0A1B6MBF9 A0A0P5C6B6 A0A1A9VV94 A0A1B0B115 A0A0A9WDU0 B4JJP0 A0A0L7QL39 A0A0P5D7T9 A0A0P5C3V9 E0VWU8 A0A1A9Z9R4 A0A0P5GG49 A0A224XI17 A0A0P5C684 A0A2R7WCH9 A0A1A9WVE6 A0A0P5C405 A0A0V0G688 B4NBZ3 A0A1B0FIT2 T1PAB0 A0A0P5CDF4 T1IAM2 A0A069DRQ9 A0A0P4VHG5 A0A161MZC9 A0A0P5CG43 A0A0N8AV13 A0A1I8Q9L0 A0A0P5C5K1 A0A023FCW0 A0A3S3PUL4 T1KPN5 A0A0P5C5K7 A0A0P5PL76 A0A0P4YJ31 A0A0P6EU95 A0A0P5EPL9 X2JFR0 A0A0K2UDU8 A0A0P5C5P6 A0A0P5CGY2 A0A0N8AKN4 B4I718 Q8MZ13
A0A182LYX9 A0A226EN38 A0A088AUZ8 A0A1J1HFT6 A0A0P4W6L2 A0A182KBP2 V5GSQ7 A0A0P4W219 Q16P12 A0A023EPG9 A0A182XWM5 A0A2M4ANQ3 A0A182SLH3 Q7PCP9 A0A182HJL4 A0A182XKD0 A0A336K6L8 A0A182G0W2 A0A182P8N4 A0A2M4CMD3 F5HLR6 A0A1Q3G4I8 A0A1L8E079 A0A1L8E0D3 A0A0K8U3Q4 A0A1L8E0G5 A0A0A1X2W7 A0A182F7V9 W5JCU4 A0A0K8WIH9 A0A034WG10 A0A1B6BXA4 W8C0I7 A0A182NSR8 A0A067QRR3 F5HLR5 A0A182U4Y2 A0A182VY90 N6T1T2 A0A1B6J1H7 A0A1B6HNR5 J3JXA8 B4L4F0 K7IZ49 A0A084VUZ2 B4M2Q4 A0A0L0BQ17 A0A0A9WAT4 A0A1B6MBF9 A0A0P5C6B6 A0A1A9VV94 A0A1B0B115 A0A0A9WDU0 B4JJP0 A0A0L7QL39 A0A0P5D7T9 A0A0P5C3V9 E0VWU8 A0A1A9Z9R4 A0A0P5GG49 A0A224XI17 A0A0P5C684 A0A2R7WCH9 A0A1A9WVE6 A0A0P5C405 A0A0V0G688 B4NBZ3 A0A1B0FIT2 T1PAB0 A0A0P5CDF4 T1IAM2 A0A069DRQ9 A0A0P4VHG5 A0A161MZC9 A0A0P5CG43 A0A0N8AV13 A0A1I8Q9L0 A0A0P5C5K1 A0A023FCW0 A0A3S3PUL4 T1KPN5 A0A0P5C5K7 A0A0P5PL76 A0A0P4YJ31 A0A0P6EU95 A0A0P5EPL9 X2JFR0 A0A0K2UDU8 A0A0P5C5P6 A0A0P5CGY2 A0A0N8AKN4 B4I718 Q8MZ13
PDB
4FVF
E-value=5.48656e-40,
Score=412
Ontologies
GO
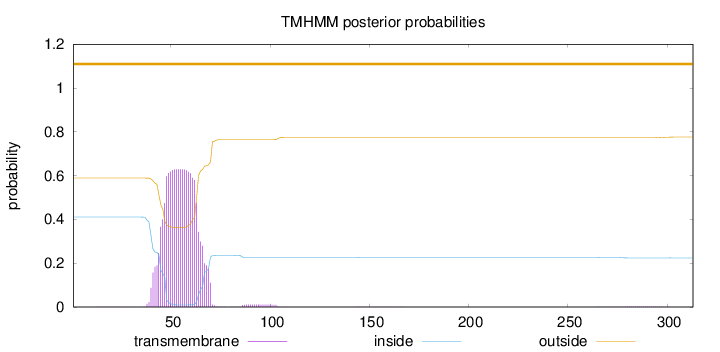
Topology
Length:
313
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
13.72507
Exp number, first 60 AAs:
10.22025
Total prob of N-in:
0.41111
POSSIBLE N-term signal
sequence
outside
1 - 313
Population Genetic Test Statistics
Pi
269.539655
Theta
200.451174
Tajima's D
1.630055
CLR
0.324628
CSRT
0.813459327033648
Interpretation
Uncertain
