Gene
KWMTBOMO12965 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000447
Annotation
N-ethylmaleimide_sensitive_fusion_protein_[Danaus_plexippus]
Full name
Vesicle-fusing ATPase 2
Alternative Name
N-ethylmaleimide-sensitive fusion protein 2
Vesicular-fusion protein NSF2
dNsf-2
Vesicular-fusion protein NSF2
dNsf-2
Location in the cell
Nuclear Reliability : 2.427
Sequence
CDS
ATGTCTTCAATGCGAATGAAAGCAGCCAAATGTCCATCTGATGAGCTTGCCATCACCAACTGTGCTCTAATACATCAAGATGACTTTCCAAGTGACATCAAGCATATTGAAGTATCTACAGGACCATCTCAACACTTTGTGTTCAGCATTCGGTTCTACAATGGAGTTGACCGTGGCACTGTTGGATTTTCTGCCCCTCAGCGAAAATGGGCTACCCTCTCTATCGGTCAAACCATTGACGTGAAGCCTTTCAAACCCTCTAGTGCTGAATGCCTTTGCAGCGTAACGTTGGAAGCCGATTTCATGATGAAGAAAACAACATCAACGGATCCATATGATTCGGAACAAATGGCCAGAGATTTCCTTATTCAGTTCGCCAATCAAGTTTTTACTGTCGGACAACAGCTGGCTTTCTTGTTCCAAGAGAAGAAAGTCTTGTCGTTGATCGTAAAAAATTTGGAAGCCGTCGATGTGCAAGCTTTAGCGGCCGGAGCCAACGCCGTGCCCCGCCGGGTGCGGATGGGCCGGCTGCTGCCCGACGCCTCGGTGCAGTTCGACAAGGCGGAGAATTCGTCTCTGAACCTAATCGGGAAGGCTAAAGGAAAGCAACCGCGACAGTCGATCATCAATCCGGACTGGGACTTCGGCAAGATGGGCATCGGCGGTCTCGACAACGAGTTCAACGCGATCTTCCGTCGCGCGTTTGCTTCCCGAGTGTTCCCCCCGGAGGTCGTCGAGCAACTAGGCTGCAAGCACGTGAAGGGAATCTTGCTCTACGGTCCGCCCGGCACCGGCAAGACTTTGATGGCCCGACAGATCGGGAAGATGCTAAACGCCCGCGAACCGAAGATCGTCAACGGCCCACAGATATTGGACAAATACGTTGGCGAGAGCGAGGCCAACATTCGACGTTTATTTGCCGACGCCGAGGAAGAAGAGAAGCGATGCGGAGCGAACAGCGGGCTTCACATCATAATATTCGATGAAATCGACGCGATCTGCAAGGCCAGGGGCTCCGTCGGCGGCAACACCGGCGTCCACGACACCGTCGTCAATCAGCTCCTCTCCAAGATCGACGGTGTGGATCAACTGAACAATATCCTGGTGATCGGTATGACGAACCGGCGCGACATGATCGACGAGGCCCTGCTCCGACCCGGCCGTCTCGAGGTGCAGATGGAGATCGGGCTTCCCGACGAGAAGGGGCGCGTCCAGATATTGAACATTCACACCAAGCGCATGCGCGAGTATAAGAAAATCGCCGAAGACGTCGACTCTATGGAGTTGGCAACGTTAACGAAGAACTTCTCTGGGGCCGAACTGGAGGGTTTGGTCCGAGCTGCCCAGTCGACCGCCATGAACAGACTCATCAAGGCGTCGAGCAAAGTCGAAGTCGATCCCGAAGCCATGGAGAAACTGATGGTCGAACGTACCGACTTCTTGCACGCCTTGGAGAATGATATCAAACCGGCCTTCGGTACATCCGCCGAAGCGCTCGAGCACTTCTTGGCTCGCGGCATCGTCAACTGGGGCTCTCCTGTTTCTTCTATATTTGAAGATGGACAGTTGTATATTCAACAGGCGCGGGCGACCGAAGCCAGCGGTCTGGTGTCGGTGCTGTTGGAGGGCCCCCCGAACAGCGGCAAGACGGCCCTCGCCGCGCAACTGGCCAAGCTGTCGGACTTCCCGTTCGTGAAGGTTTGCTCGCCCGAAGACATGGTCGGCTTCACGGAGACAGCTAAATGCTTGCAGATTAGGAAGTATTTCGACGACGCGTACCGCTCGAGCCTGTCGTGCATCCTGGTCGACAACATCGAGCGGCTGCTGGACTACGGACCGATCGGGCCGCGCTACTCCAACCTGACGCTGCAGGCGCTGCTCGTTCTCCTGAAGAAGCAGCCGCCCAAGGGACGAAAACTGCTCATCTTGTGCACGAGCAGCCGAAGGCAAGTGTTGGAAGACATGGAAATCCTGTCTGCGTTCACCGGAGTGCTCCACGTGCCGAACCTGTCGCAGCCCGAGCACCTGCTGTCGGTGCTCGAGGAGAGCGAGGCCTTCTCCAAGCGCGACCTGCAGAAGATACAGAGCGACCTGCGAGGAGCCAAAATCTTCATCGGTATAAAGAAGCTGCTCGCTCTAATCGACATGGTGAAGCAGACCGACGAAGAATACAGAGTGTTCAAGTTCCTCACGAAGTTGCAGGAGGAGGGCTGCGGATTACACGCGCATACCACGTCGCCCACGACCGACCACACCGCGACCCTTTGTACGTCCAGAACTCACGGCGCCAAACTAACCGAGTGCGCACCTTGCACTTCAGACCTCCGCAGCTATGAAATAGACTACGATGACGTCAGCGAATGCACCGACTCTCCCTTAGACATCGGCTATCACAAAAAAGAAATGGCCGCCAAAAGGACCGAGATATCGCCCGCCAAAATGCCGCTGATGCGAATTCGAAGCGTCGAGATTGTATTTGAAGACGAAATGTCGACTTGCACCGACACCGGGCTCCTGGACCGCAGCGTGGAACTCGTCGTCTCGGACAGTAGCGATGATTCCACTAAATGCGCGGACGGTATCGAGCCTCTACCGCCCGAGAAAGATTGCGACTCTGTGACGCTCAACGCTGACGGCGACTTGTCGGAGGACGACTCATCATCGTCGCTGAACAAACACGAAATACCTGAAATCGTTATTCCACAGCCGACGGCTCCGGCACACGCGACCCTTCTAGTTCGAACAGAGAACGTAACGTCGCGGACGGATAATTTGGCGATGTCGCCAAGTTTCTCTGAAATGGCAGACTCCGGTCGATACACAGAGGAAGCTAGTTCATCCCATATAGACATCGTTGCGATGTCTTACGAAGACGCATTGCTTTCAAACTCCTTTGACGTTATCAATTTAGAGGAACTTCCGGCCGCGACTCGCTCTTACGATTCGGACTCAAGCTTGAAACCAGAGACTCCGAGATTATTGGAATACATTGACAGTAACAGTGACAGCTCAGACACGGAAACGTGTGACTTGGACAGTCGAACGCATCCTCTGACTCCTCTCAACTTCATCTTGTCCGATGACGACCACACGGTGTGCGACAGTTCCGATTTGCAGTACGACTCTTTTGTAGATATGGGGAAGTTTTAA
Protein
MSSMRMKAAKCPSDELAITNCALIHQDDFPSDIKHIEVSTGPSQHFVFSIRFYNGVDRGTVGFSAPQRKWATLSIGQTIDVKPFKPSSAECLCSVTLEADFMMKKTTSTDPYDSEQMARDFLIQFANQVFTVGQQLAFLFQEKKVLSLIVKNLEAVDVQALAAGANAVPRRVRMGRLLPDASVQFDKAENSSLNLIGKAKGKQPRQSIINPDWDFGKMGIGGLDNEFNAIFRRAFASRVFPPEVVEQLGCKHVKGILLYGPPGTGKTLMARQIGKMLNAREPKIVNGPQILDKYVGESEANIRRLFADAEEEEKRCGANSGLHIIIFDEIDAICKARGSVGGNTGVHDTVVNQLLSKIDGVDQLNNILVIGMTNRRDMIDEALLRPGRLEVQMEIGLPDEKGRVQILNIHTKRMREYKKIAEDVDSMELATLTKNFSGAELEGLVRAAQSTAMNRLIKASSKVEVDPEAMEKLMVERTDFLHALENDIKPAFGTSAEALEHFLARGIVNWGSPVSSIFEDGQLYIQQARATEASGLVSVLLEGPPNSGKTALAAQLAKLSDFPFVKVCSPEDMVGFTETAKCLQIRKYFDDAYRSSLSCILVDNIERLLDYGPIGPRYSNLTLQALLVLLKKQPPKGRKLLILCTSSRRQVLEDMEILSAFTGVLHVPNLSQPEHLLSVLEESEAFSKRDLQKIQSDLRGAKIFIGIKKLLALIDMVKQTDEEYRVFKFLTKLQEEGCGLHAHTTSPTTDHTATLCTSRTHGAKLTECAPCTSDLRSYEIDYDDVSECTDSPLDIGYHKKEMAAKRTEISPAKMPLMRIRSVEIVFEDEMSTCTDTGLLDRSVELVVSDSSDDSTKCADGIEPLPPEKDCDSVTLNADGDLSEDDSSSSLNKHEIPEIVIPQPTAPAHATLLVRTENVTSRTDNLAMSPSFSEMADSGRYTEEASSSHIDIVAMSYEDALLSNSFDVINLEELPAATRSYDSDSSLKPETPRLLEYIDSNSDSSDTETCDLDSRTHPLTPLNFILSDDDHTVCDSSDLQYDSFVDMGKF
Summary
Description
Required for vesicle-mediated transport. Catalyzes the fusion of transport vesicles within the Golgi cisternae. Is also required for transport from the endoplasmic reticulum to the Golgi stack. Seems to function as a fusion protein required for the delivery of cargo proteins to all compartments of the Golgi stack independent of vesicle origin (By similarity).
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Subunit
Homohexamer.
Similarity
Belongs to the AAA ATPase family.
Keywords
ATP-binding
Complete proteome
Cytoplasm
ER-Golgi transport
Hydrolase
Magnesium
Metal-binding
Nucleotide-binding
Protein transport
Reference proteome
Repeat
Transport
Feature
chain Vesicle-fusing ATPase 2
Uniprot
A0A212F4N3
A0A2A4JAG1
A0A1P8W8N0
Q9U4Y6
A0A2A4J9E7
A0A2H1VZQ1
+ More
A0A194Q822 A0A194QWW7 A0A1B6G249 A0A1B6MSH2 A0A2J7QCI0 A0A0M8ZZU6 A0A1B6C6V7 A0A067RCN5 A0A088A494 V9IKG1 A0A2A3E1X9 A0A1B0CWA3 A0A1Y1KMW4 A0A232F624 A0A1L8DW68 A0A0C9QG91 A0A0K8TTK0 E2C1M2 A0A158NK70 A0A0C9RWV1 A0A195BAX8 A0A195FBP9 F4W8G6 A0A151X6W0 Q17KM8 A0A195CZY0 A0A182GIC0 Q6EJD3 A0A0L7QQB5 A0A1Q3G3L1 A0A195EHT9 A0A0A1XP20 A0A1J1ILN9 A0A0L0C0M2 A0A026WU40 A0A0K8U5G2 A0A034VKS2 E2B0I4 W8AZC2 U5EWA2 B4MA92 B4L596 A0A1I8QDC7 B4NPN2 A0A3B0J6J7 T1PF76 B3NWQ9 B4Q231 A0A1A9WAQ9 D6X0K4 B4JMJ4 A0A0M3QY10 B3MS56 A0A0M5JE01 A0A1W4V7U5 B5DKI8 B4H3A0 A0A1B0FLT1 A0A1W4V6F2 B3LVS7 A0A182W7Y6 B4PQJ8 A0A182YDX3 B3P3Z3 A0A0B4LH53 P54351 T1IP97 A0A336LSI7 E0VAZ5 A0A146LHD2 B4HF87 A0A164W5F2 A0A0A9WYG7
A0A194Q822 A0A194QWW7 A0A1B6G249 A0A1B6MSH2 A0A2J7QCI0 A0A0M8ZZU6 A0A1B6C6V7 A0A067RCN5 A0A088A494 V9IKG1 A0A2A3E1X9 A0A1B0CWA3 A0A1Y1KMW4 A0A232F624 A0A1L8DW68 A0A0C9QG91 A0A0K8TTK0 E2C1M2 A0A158NK70 A0A0C9RWV1 A0A195BAX8 A0A195FBP9 F4W8G6 A0A151X6W0 Q17KM8 A0A195CZY0 A0A182GIC0 Q6EJD3 A0A0L7QQB5 A0A1Q3G3L1 A0A195EHT9 A0A0A1XP20 A0A1J1ILN9 A0A0L0C0M2 A0A026WU40 A0A0K8U5G2 A0A034VKS2 E2B0I4 W8AZC2 U5EWA2 B4MA92 B4L596 A0A1I8QDC7 B4NPN2 A0A3B0J6J7 T1PF76 B3NWQ9 B4Q231 A0A1A9WAQ9 D6X0K4 B4JMJ4 A0A0M3QY10 B3MS56 A0A0M5JE01 A0A1W4V7U5 B5DKI8 B4H3A0 A0A1B0FLT1 A0A1W4V6F2 B3LVS7 A0A182W7Y6 B4PQJ8 A0A182YDX3 B3P3Z3 A0A0B4LH53 P54351 T1IP97 A0A336LSI7 E0VAZ5 A0A146LHD2 B4HF87 A0A164W5F2 A0A0A9WYG7
EC Number
3.6.4.6
Pubmed
22118469
10580154
26354079
24845553
28004739
28648823
+ More
26369729 20798317 21347285 21719571 17510324 26483478 25830018 26108605 24508170 25348373 24495485 17994087 25315136 17550304 18362917 19820115 18057021 15632085 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 7642522 7624376 20566863 26823975 25401762
26369729 20798317 21347285 21719571 17510324 26483478 25830018 26108605 24508170 25348373 24495485 17994087 25315136 17550304 18362917 19820115 18057021 15632085 25244985 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 7642522 7624376 20566863 26823975 25401762
EMBL
AGBW02010334
OWR48697.1
NWSH01002266
PCG68746.1
KY548397
APZ89680.1
+ More
AF118384 AAF18300.1 PCG68747.1 ODYU01005445 SOQ46310.1 KQ459324 KPJ01539.1 KQ461073 KPJ09799.1 GECZ01017003 GECZ01013252 JAS52766.1 JAS56517.1 GEBQ01001090 JAT38887.1 NEVH01016289 PNF26292.1 KQ435789 KOX74480.1 GEDC01030689 GEDC01028091 GEDC01023926 GEDC01023325 GEDC01013279 JAS06609.1 JAS09207.1 JAS13372.1 JAS13973.1 JAS24019.1 KK852788 KDR16531.1 JR049505 AEY60986.1 KZ288470 PBC25332.1 AJWK01032124 AJWK01032125 AJWK01032126 GEZM01078395 GEZM01078394 JAV62742.1 NNAY01000881 OXU26085.1 GFDF01003394 JAV10690.1 GBYB01013543 JAG83310.1 GDAI01000338 JAI17265.1 GL451937 EFN78198.1 ADTU01018723 GBYB01013545 JAG83312.1 KQ976537 KYM81375.1 KQ981693 KYN37637.1 GL887908 EGI69457.1 KQ982472 KYQ56092.1 CH477222 EAT47276.1 KQ977110 KYN05689.1 JXUM01065739 JXUM01065740 KQ562363 KXJ76058.1 AY314995 AAQ83118.1 KQ414786 KOC60817.1 GFDL01000653 JAV34392.1 KQ978881 KYN27818.1 GBXI01001600 JAD12692.1 CVRI01000054 CRK99990.1 JRES01001065 KNC25860.1 KK107105 EZA59590.1 GDHF01030491 JAI21823.1 GAKP01015041 JAC43911.1 GL444608 EFN60826.1 GAMC01012390 JAB94165.1 GANO01001513 JAB58358.1 CH940655 EDW66151.1 CH933811 EDW06355.1 CH964291 EDW86472.1 OUUW01000003 SPP77784.1 KA646815 AFP61444.1 CH954180 EDV47221.1 CM000162 EDX01552.1 KQ971371 EFA10100.1 CH916371 EDV91937.1 CP012526 ALC46809.1 CH902622 EDV34611.1 KPU75214.1 CP012528 ALC49856.1 CH379063 EDY72037.1 CH479205 EDW30793.1 CCAG010020037 CH902617 EDV43701.2 CM000160 EDW97294.1 CH954181 EDV49167.1 AE014297 AHN57317.1 AHN57318.1 U30502 BT023784 U28836 JH431251 UFQT01000155 SSX20982.1 DS235019 EEB10551.1 GDHC01012273 JAQ06356.1 CH480815 EDW42261.1 LRGB01001348 KZS12965.1 GBHO01030815 GBHO01002606 GBHO01002601 JAG12789.1 JAG40998.1 JAG41003.1
AF118384 AAF18300.1 PCG68747.1 ODYU01005445 SOQ46310.1 KQ459324 KPJ01539.1 KQ461073 KPJ09799.1 GECZ01017003 GECZ01013252 JAS52766.1 JAS56517.1 GEBQ01001090 JAT38887.1 NEVH01016289 PNF26292.1 KQ435789 KOX74480.1 GEDC01030689 GEDC01028091 GEDC01023926 GEDC01023325 GEDC01013279 JAS06609.1 JAS09207.1 JAS13372.1 JAS13973.1 JAS24019.1 KK852788 KDR16531.1 JR049505 AEY60986.1 KZ288470 PBC25332.1 AJWK01032124 AJWK01032125 AJWK01032126 GEZM01078395 GEZM01078394 JAV62742.1 NNAY01000881 OXU26085.1 GFDF01003394 JAV10690.1 GBYB01013543 JAG83310.1 GDAI01000338 JAI17265.1 GL451937 EFN78198.1 ADTU01018723 GBYB01013545 JAG83312.1 KQ976537 KYM81375.1 KQ981693 KYN37637.1 GL887908 EGI69457.1 KQ982472 KYQ56092.1 CH477222 EAT47276.1 KQ977110 KYN05689.1 JXUM01065739 JXUM01065740 KQ562363 KXJ76058.1 AY314995 AAQ83118.1 KQ414786 KOC60817.1 GFDL01000653 JAV34392.1 KQ978881 KYN27818.1 GBXI01001600 JAD12692.1 CVRI01000054 CRK99990.1 JRES01001065 KNC25860.1 KK107105 EZA59590.1 GDHF01030491 JAI21823.1 GAKP01015041 JAC43911.1 GL444608 EFN60826.1 GAMC01012390 JAB94165.1 GANO01001513 JAB58358.1 CH940655 EDW66151.1 CH933811 EDW06355.1 CH964291 EDW86472.1 OUUW01000003 SPP77784.1 KA646815 AFP61444.1 CH954180 EDV47221.1 CM000162 EDX01552.1 KQ971371 EFA10100.1 CH916371 EDV91937.1 CP012526 ALC46809.1 CH902622 EDV34611.1 KPU75214.1 CP012528 ALC49856.1 CH379063 EDY72037.1 CH479205 EDW30793.1 CCAG010020037 CH902617 EDV43701.2 CM000160 EDW97294.1 CH954181 EDV49167.1 AE014297 AHN57317.1 AHN57318.1 U30502 BT023784 U28836 JH431251 UFQT01000155 SSX20982.1 DS235019 EEB10551.1 GDHC01012273 JAQ06356.1 CH480815 EDW42261.1 LRGB01001348 KZS12965.1 GBHO01030815 GBHO01002606 GBHO01002601 JAG12789.1 JAG40998.1 JAG41003.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000053240
UP000235965
UP000053105
+ More
UP000027135 UP000005203 UP000242457 UP000092461 UP000215335 UP000008237 UP000005205 UP000078540 UP000078541 UP000007755 UP000075809 UP000008820 UP000078542 UP000069940 UP000249989 UP000053825 UP000078492 UP000183832 UP000037069 UP000053097 UP000000311 UP000008792 UP000009192 UP000095300 UP000007798 UP000268350 UP000095301 UP000008711 UP000002282 UP000091820 UP000007266 UP000001070 UP000092553 UP000007801 UP000192221 UP000001819 UP000008744 UP000092444 UP000075920 UP000076408 UP000000803 UP000009046 UP000001292 UP000076858
UP000027135 UP000005203 UP000242457 UP000092461 UP000215335 UP000008237 UP000005205 UP000078540 UP000078541 UP000007755 UP000075809 UP000008820 UP000078542 UP000069940 UP000249989 UP000053825 UP000078492 UP000183832 UP000037069 UP000053097 UP000000311 UP000008792 UP000009192 UP000095300 UP000007798 UP000268350 UP000095301 UP000008711 UP000002282 UP000091820 UP000007266 UP000001070 UP000092553 UP000007801 UP000192221 UP000001819 UP000008744 UP000092444 UP000075920 UP000076408 UP000000803 UP000009046 UP000001292 UP000076858
PRIDE
Interpro
ProteinModelPortal
A0A212F4N3
A0A2A4JAG1
A0A1P8W8N0
Q9U4Y6
A0A2A4J9E7
A0A2H1VZQ1
+ More
A0A194Q822 A0A194QWW7 A0A1B6G249 A0A1B6MSH2 A0A2J7QCI0 A0A0M8ZZU6 A0A1B6C6V7 A0A067RCN5 A0A088A494 V9IKG1 A0A2A3E1X9 A0A1B0CWA3 A0A1Y1KMW4 A0A232F624 A0A1L8DW68 A0A0C9QG91 A0A0K8TTK0 E2C1M2 A0A158NK70 A0A0C9RWV1 A0A195BAX8 A0A195FBP9 F4W8G6 A0A151X6W0 Q17KM8 A0A195CZY0 A0A182GIC0 Q6EJD3 A0A0L7QQB5 A0A1Q3G3L1 A0A195EHT9 A0A0A1XP20 A0A1J1ILN9 A0A0L0C0M2 A0A026WU40 A0A0K8U5G2 A0A034VKS2 E2B0I4 W8AZC2 U5EWA2 B4MA92 B4L596 A0A1I8QDC7 B4NPN2 A0A3B0J6J7 T1PF76 B3NWQ9 B4Q231 A0A1A9WAQ9 D6X0K4 B4JMJ4 A0A0M3QY10 B3MS56 A0A0M5JE01 A0A1W4V7U5 B5DKI8 B4H3A0 A0A1B0FLT1 A0A1W4V6F2 B3LVS7 A0A182W7Y6 B4PQJ8 A0A182YDX3 B3P3Z3 A0A0B4LH53 P54351 T1IP97 A0A336LSI7 E0VAZ5 A0A146LHD2 B4HF87 A0A164W5F2 A0A0A9WYG7
A0A194Q822 A0A194QWW7 A0A1B6G249 A0A1B6MSH2 A0A2J7QCI0 A0A0M8ZZU6 A0A1B6C6V7 A0A067RCN5 A0A088A494 V9IKG1 A0A2A3E1X9 A0A1B0CWA3 A0A1Y1KMW4 A0A232F624 A0A1L8DW68 A0A0C9QG91 A0A0K8TTK0 E2C1M2 A0A158NK70 A0A0C9RWV1 A0A195BAX8 A0A195FBP9 F4W8G6 A0A151X6W0 Q17KM8 A0A195CZY0 A0A182GIC0 Q6EJD3 A0A0L7QQB5 A0A1Q3G3L1 A0A195EHT9 A0A0A1XP20 A0A1J1ILN9 A0A0L0C0M2 A0A026WU40 A0A0K8U5G2 A0A034VKS2 E2B0I4 W8AZC2 U5EWA2 B4MA92 B4L596 A0A1I8QDC7 B4NPN2 A0A3B0J6J7 T1PF76 B3NWQ9 B4Q231 A0A1A9WAQ9 D6X0K4 B4JMJ4 A0A0M3QY10 B3MS56 A0A0M5JE01 A0A1W4V7U5 B5DKI8 B4H3A0 A0A1B0FLT1 A0A1W4V6F2 B3LVS7 A0A182W7Y6 B4PQJ8 A0A182YDX3 B3P3Z3 A0A0B4LH53 P54351 T1IP97 A0A336LSI7 E0VAZ5 A0A146LHD2 B4HF87 A0A164W5F2 A0A0A9WYG7
PDB
3J99
E-value=0,
Score=2406
Ontologies
GO
GO:0035494
GO:0005524
GO:0016887
GO:0070050
GO:0008021
GO:0007274
GO:0090160
GO:0048172
GO:0043195
GO:0005829
GO:1900073
GO:0007030
GO:0006914
GO:0098527
GO:0031629
GO:0005795
GO:0043001
GO:0048211
GO:0008582
GO:0060439
GO:0007269
GO:0016082
GO:0046872
GO:0007424
GO:0016192
GO:0005737
GO:0098793
GO:0035002
GO:0007430
GO:0015031
GO:0008270
GO:0003700
GO:0043565
GO:0007155
GO:0005515
GO:0008237
GO:0008152
GO:0016491
GO:0015930
PANTHER
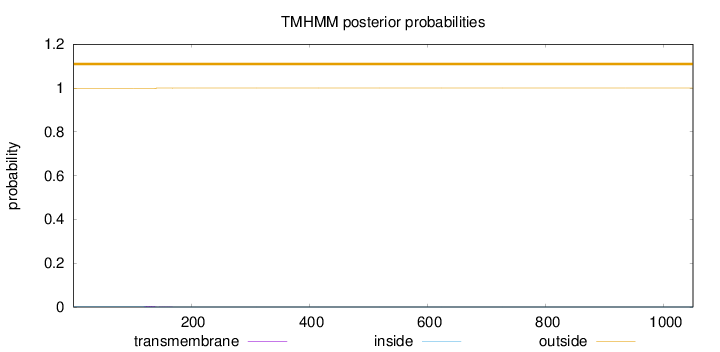
Topology
Subcellular location
Cytoplasm
Length:
1049
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0538700000000001
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.00282
outside
1 - 1049
Population Genetic Test Statistics
Pi
213.975098
Theta
150.621873
Tajima's D
1.81116
CLR
0.283492
CSRT
0.853557322133893
Interpretation
Uncertain
