Gene
KWMTBOMO12964
Annotation
PREDICTED:_transcription_elongation_factor_1_homolog_[Plutella_xylostella]
Full name
Transcription elongation factor 1 homolog
Location in the cell
Nuclear Reliability : 3.576
Sequence
CDS
ATGGGACGTCGAAAATCAAAAAGGAAGCCACCACCAAAACGAAAGGCTACCGAGGCACTGGATCAGCAATTTAACTGTCCGTTTTGCAATCATGAAAAATCCTGCGAAGTAAAAATGGATCGCGCAAGGAATACTGCGCGAATACAGTGTCGAGTTTGTTTAGAAGATTTCCAAACAACAACAAATGTTTTGTCTGAGCCAATAGATGTATATAATGACTGGGTGGATGCTTGTGAAAGTGCGAATTAA
Protein
MGRRKSKRKPPPKRKATEALDQQFNCPFCNHEKSCEVKMDRARNTARIQCRVCLEDFQTTTNVLSEPIDVYNDWVDACESAN
Summary
Description
Transcription elongation factor implicated in the maintenance of proper chromatin structure in actively transcribed regions.
Similarity
Belongs to the ELOF1 family.
Keywords
Metal-binding
Nucleus
Transcription
Transcription regulation
Zinc
Zinc-finger
Complete proteome
Reference proteome
Feature
chain Transcription elongation factor 1 homolog
Uniprot
A0A194R1S4
A0A194QDK6
A0A2H1VZP1
S4Q0B2
A0A212F4M9
Q9U501
+ More
D6X363 A0A336LJA0 A0A232EFM3 K7J4X2 A0A1L8DHP5 U5EZF0 B4IV40 A0A224Y464 A0A0V0G970 A0A023F9D0 A0A069DVA1 A0A2A3EMJ4 A0A088AKB0 A0A2P8YQ77 A0A1W4VH03 B4MGG1 B4K4E8 B5DSA5 B4GE72 A0A0M4ECL6 B3MTK8 B4JHQ2 B3NJ89 B4NV21 B4INN1 L7EEF9 Q8MQI6 D3TLC9 A0A1A9XEY4 A0A1B0ARE6 A0A1B0ADA5 A0A1A9W8D4 A0A1B0EZI1 A0A1B0CEK0 A0A0P4W1Q8 R4FMJ2 Q0IFZ5 A0A023EE27 A0A0L7R030 A0A1Q3F5H5 B0WEX0 A0A1B0FCQ6 B4NA07 A0A0J7NAY3 A0A026X1S1 A0A154PGE5 A0A195CT75 A0A1A9VQ55 T1H732 A0A0K8TP13 R4UJU5 A0A0L0CCD2 E9IRQ5 A0A0N0BF85 A0A151X763 T1JJN4 F4WZ99 A0A195F7Q9 A0A158P1I6 N6U4Q3 A0A1W4WPC6 A0A151IZ15 A0A182PTZ8 A0A154PL55 A0A2J7PHF4 A0A067QXF0 A0A1S3DTG5 J9KPW7 A0A087UK15 A0A1I8M637 A0A182YEV5 A0A182LZ49 A0A182S6S0 A0A182V886 A0A182UK62 A0A182KXY1 Q7PH52 A0A182R8D6 A0A182WVF1 E2AFU8 A0A1Y9HA44 A0A182W2Z8 A0A084WR31 A0A3S3SPR9 A0A1L8EDQ6 A0A2M3ZDH0 A0A2M4AYZ2 A0A2M4C506 W5JGJ2 A0A182FX91 A0A0L7QL26 A0A2R7WD39 E0VKM1 A0A293LKJ5
D6X363 A0A336LJA0 A0A232EFM3 K7J4X2 A0A1L8DHP5 U5EZF0 B4IV40 A0A224Y464 A0A0V0G970 A0A023F9D0 A0A069DVA1 A0A2A3EMJ4 A0A088AKB0 A0A2P8YQ77 A0A1W4VH03 B4MGG1 B4K4E8 B5DSA5 B4GE72 A0A0M4ECL6 B3MTK8 B4JHQ2 B3NJ89 B4NV21 B4INN1 L7EEF9 Q8MQI6 D3TLC9 A0A1A9XEY4 A0A1B0ARE6 A0A1B0ADA5 A0A1A9W8D4 A0A1B0EZI1 A0A1B0CEK0 A0A0P4W1Q8 R4FMJ2 Q0IFZ5 A0A023EE27 A0A0L7R030 A0A1Q3F5H5 B0WEX0 A0A1B0FCQ6 B4NA07 A0A0J7NAY3 A0A026X1S1 A0A154PGE5 A0A195CT75 A0A1A9VQ55 T1H732 A0A0K8TP13 R4UJU5 A0A0L0CCD2 E9IRQ5 A0A0N0BF85 A0A151X763 T1JJN4 F4WZ99 A0A195F7Q9 A0A158P1I6 N6U4Q3 A0A1W4WPC6 A0A151IZ15 A0A182PTZ8 A0A154PL55 A0A2J7PHF4 A0A067QXF0 A0A1S3DTG5 J9KPW7 A0A087UK15 A0A1I8M637 A0A182YEV5 A0A182LZ49 A0A182S6S0 A0A182V886 A0A182UK62 A0A182KXY1 Q7PH52 A0A182R8D6 A0A182WVF1 E2AFU8 A0A1Y9HA44 A0A182W2Z8 A0A084WR31 A0A3S3SPR9 A0A1L8EDQ6 A0A2M3ZDH0 A0A2M4AYZ2 A0A2M4C506 W5JGJ2 A0A182FX91 A0A0L7QL26 A0A2R7WD39 E0VKM1 A0A293LKJ5
Pubmed
26354079
23622113
22118469
10620045
18362917
19820115
+ More
28648823 20075255 17994087 17550304 25474469 26334808 29403074 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 20353571 27129103 17510324 24945155 26483478 24508170 30249741 26369729 26108605 21282665 21719571 21347285 23537049 24845553 25315136 25244985 20966253 12364791 14747013 17210077 20798317 24438588 20920257 23761445 20566863
28648823 20075255 17994087 17550304 25474469 26334808 29403074 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 20353571 27129103 17510324 24945155 26483478 24508170 30249741 26369729 26108605 21282665 21719571 21347285 23537049 24845553 25315136 25244985 20966253 12364791 14747013 17210077 20798317 24438588 20920257 23761445 20566863
EMBL
KQ461073
KPJ09801.1
KQ459324
KPJ01541.1
ODYU01005445
SOQ46311.1
+ More
GAIX01000093 JAA92467.1 AGBW02010334 OWR48695.1 AF117587 KQ971372 EFA10805.1 UFQS01005063 UFQT01001174 UFQT01005063 SSX16687.1 SSX29298.1 NNAY01004967 OXU17160.1 GFDF01008095 JAV05989.1 GANO01000916 JAB58955.1 CH892605 EDW99604.1 GFTR01000943 JAW15483.1 GECL01001667 JAP04457.1 GBBI01000606 JAC18106.1 GBGD01003655 JAC85234.1 KZ288217 PBC32379.1 PYGN01000437 PSN46408.1 CH940675 EDW58211.1 CH933806 EDW13900.1 CH475481 EDY71453.1 CH479182 EDW33907.1 CP012526 ALC45416.1 CH902623 EDV30139.1 CH916369 EDV93891.1 CH954178 EDV52872.1 CH985487 EDX15896.1 CH676488 EDW51374.1 KX531586 AE014296 ANY27396.1 ELP57411.1 AY129458 EZ422231 ADD18507.1 JXJN01002381 AJVK01007413 AJWK01008941 GDKW01001012 JAI55583.1 ACPB03021649 GAHY01000973 JAA76537.1 CH477281 EAT44929.1 JXUM01015655 JXUM01126288 GAPW01006719 KQ567050 KQ560436 JAC06879.1 KXJ69649.1 KXJ82430.1 KQ414672 KOC64210.1 GFDL01012233 JAV22812.1 DS231912 EDS25828.1 CCAG010006641 CH964232 EDW81762.1 LBMM01007398 KMQ89765.1 KK107024 QOIP01000004 EZA62217.1 RLU23910.1 KQ434889 KZC10260.1 KQ977305 KYN03722.1 GDAI01001712 JAI15891.1 KC571850 AGM32349.1 JRES01000577 KNC30098.1 GL765283 EFZ16624.1 KQ435808 KOX72992.1 KQ982450 KYQ56212.1 JH431929 GL888470 EGI60488.1 KQ981756 KYN36099.1 ADTU01006685 APGK01042895 KB741009 KB632250 ENN75601.1 ERL90595.1 KQ980735 KYN13765.1 KQ434954 KZC12611.1 NEVH01025138 PNF15760.1 KK852858 KDR14863.1 ABLF02039777 KK120184 KFM77704.1 AXCM01000016 AAAB01008888 EAA44682.1 GL439158 EFN67697.1 AXCN02001184 ATLV01025911 KE525402 KFB52675.1 NCKU01000107 RWS17217.1 GFDG01001972 JAV16827.1 GGFM01005804 MBW26555.1 GGFK01012683 MBW46004.1 GGFJ01011272 MBW60413.1 ADMH02001252 ETN63482.1 KQ414934 KOC59327.1 KK854643 PTY17576.1 DS235248 EEB13927.1 GFWV01003631 MAA28361.1
GAIX01000093 JAA92467.1 AGBW02010334 OWR48695.1 AF117587 KQ971372 EFA10805.1 UFQS01005063 UFQT01001174 UFQT01005063 SSX16687.1 SSX29298.1 NNAY01004967 OXU17160.1 GFDF01008095 JAV05989.1 GANO01000916 JAB58955.1 CH892605 EDW99604.1 GFTR01000943 JAW15483.1 GECL01001667 JAP04457.1 GBBI01000606 JAC18106.1 GBGD01003655 JAC85234.1 KZ288217 PBC32379.1 PYGN01000437 PSN46408.1 CH940675 EDW58211.1 CH933806 EDW13900.1 CH475481 EDY71453.1 CH479182 EDW33907.1 CP012526 ALC45416.1 CH902623 EDV30139.1 CH916369 EDV93891.1 CH954178 EDV52872.1 CH985487 EDX15896.1 CH676488 EDW51374.1 KX531586 AE014296 ANY27396.1 ELP57411.1 AY129458 EZ422231 ADD18507.1 JXJN01002381 AJVK01007413 AJWK01008941 GDKW01001012 JAI55583.1 ACPB03021649 GAHY01000973 JAA76537.1 CH477281 EAT44929.1 JXUM01015655 JXUM01126288 GAPW01006719 KQ567050 KQ560436 JAC06879.1 KXJ69649.1 KXJ82430.1 KQ414672 KOC64210.1 GFDL01012233 JAV22812.1 DS231912 EDS25828.1 CCAG010006641 CH964232 EDW81762.1 LBMM01007398 KMQ89765.1 KK107024 QOIP01000004 EZA62217.1 RLU23910.1 KQ434889 KZC10260.1 KQ977305 KYN03722.1 GDAI01001712 JAI15891.1 KC571850 AGM32349.1 JRES01000577 KNC30098.1 GL765283 EFZ16624.1 KQ435808 KOX72992.1 KQ982450 KYQ56212.1 JH431929 GL888470 EGI60488.1 KQ981756 KYN36099.1 ADTU01006685 APGK01042895 KB741009 KB632250 ENN75601.1 ERL90595.1 KQ980735 KYN13765.1 KQ434954 KZC12611.1 NEVH01025138 PNF15760.1 KK852858 KDR14863.1 ABLF02039777 KK120184 KFM77704.1 AXCM01000016 AAAB01008888 EAA44682.1 GL439158 EFN67697.1 AXCN02001184 ATLV01025911 KE525402 KFB52675.1 NCKU01000107 RWS17217.1 GFDG01001972 JAV16827.1 GGFM01005804 MBW26555.1 GGFK01012683 MBW46004.1 GGFJ01011272 MBW60413.1 ADMH02001252 ETN63482.1 KQ414934 KOC59327.1 KK854643 PTY17576.1 DS235248 EEB13927.1 GFWV01003631 MAA28361.1
Proteomes
UP000053240
UP000053268
UP000007151
UP000007266
UP000215335
UP000002358
+ More
UP000002282 UP000242457 UP000005203 UP000245037 UP000192221 UP000008792 UP000009192 UP000001819 UP000008744 UP000092553 UP000007801 UP000001070 UP000008711 UP000000304 UP000001292 UP000000803 UP000092443 UP000092460 UP000092445 UP000091820 UP000092462 UP000092461 UP000015103 UP000008820 UP000069940 UP000249989 UP000053825 UP000002320 UP000092444 UP000007798 UP000036403 UP000053097 UP000279307 UP000076502 UP000078542 UP000078200 UP000015102 UP000037069 UP000053105 UP000075809 UP000007755 UP000078541 UP000005205 UP000019118 UP000030742 UP000192223 UP000078492 UP000075885 UP000235965 UP000027135 UP000079169 UP000007819 UP000054359 UP000095301 UP000076408 UP000075883 UP000075901 UP000075903 UP000075902 UP000075882 UP000007062 UP000075900 UP000076407 UP000000311 UP000075886 UP000075920 UP000030765 UP000285301 UP000000673 UP000069272 UP000009046
UP000002282 UP000242457 UP000005203 UP000245037 UP000192221 UP000008792 UP000009192 UP000001819 UP000008744 UP000092553 UP000007801 UP000001070 UP000008711 UP000000304 UP000001292 UP000000803 UP000092443 UP000092460 UP000092445 UP000091820 UP000092462 UP000092461 UP000015103 UP000008820 UP000069940 UP000249989 UP000053825 UP000002320 UP000092444 UP000007798 UP000036403 UP000053097 UP000279307 UP000076502 UP000078542 UP000078200 UP000015102 UP000037069 UP000053105 UP000075809 UP000007755 UP000078541 UP000005205 UP000019118 UP000030742 UP000192223 UP000078492 UP000075885 UP000235965 UP000027135 UP000079169 UP000007819 UP000054359 UP000095301 UP000076408 UP000075883 UP000075901 UP000075903 UP000075902 UP000075882 UP000007062 UP000075900 UP000076407 UP000000311 UP000075886 UP000075920 UP000030765 UP000285301 UP000000673 UP000069272 UP000009046
PRIDE
Pfam
PF05129 Elf1
Gene 3D
ProteinModelPortal
A0A194R1S4
A0A194QDK6
A0A2H1VZP1
S4Q0B2
A0A212F4M9
Q9U501
+ More
D6X363 A0A336LJA0 A0A232EFM3 K7J4X2 A0A1L8DHP5 U5EZF0 B4IV40 A0A224Y464 A0A0V0G970 A0A023F9D0 A0A069DVA1 A0A2A3EMJ4 A0A088AKB0 A0A2P8YQ77 A0A1W4VH03 B4MGG1 B4K4E8 B5DSA5 B4GE72 A0A0M4ECL6 B3MTK8 B4JHQ2 B3NJ89 B4NV21 B4INN1 L7EEF9 Q8MQI6 D3TLC9 A0A1A9XEY4 A0A1B0ARE6 A0A1B0ADA5 A0A1A9W8D4 A0A1B0EZI1 A0A1B0CEK0 A0A0P4W1Q8 R4FMJ2 Q0IFZ5 A0A023EE27 A0A0L7R030 A0A1Q3F5H5 B0WEX0 A0A1B0FCQ6 B4NA07 A0A0J7NAY3 A0A026X1S1 A0A154PGE5 A0A195CT75 A0A1A9VQ55 T1H732 A0A0K8TP13 R4UJU5 A0A0L0CCD2 E9IRQ5 A0A0N0BF85 A0A151X763 T1JJN4 F4WZ99 A0A195F7Q9 A0A158P1I6 N6U4Q3 A0A1W4WPC6 A0A151IZ15 A0A182PTZ8 A0A154PL55 A0A2J7PHF4 A0A067QXF0 A0A1S3DTG5 J9KPW7 A0A087UK15 A0A1I8M637 A0A182YEV5 A0A182LZ49 A0A182S6S0 A0A182V886 A0A182UK62 A0A182KXY1 Q7PH52 A0A182R8D6 A0A182WVF1 E2AFU8 A0A1Y9HA44 A0A182W2Z8 A0A084WR31 A0A3S3SPR9 A0A1L8EDQ6 A0A2M3ZDH0 A0A2M4AYZ2 A0A2M4C506 W5JGJ2 A0A182FX91 A0A0L7QL26 A0A2R7WD39 E0VKM1 A0A293LKJ5
D6X363 A0A336LJA0 A0A232EFM3 K7J4X2 A0A1L8DHP5 U5EZF0 B4IV40 A0A224Y464 A0A0V0G970 A0A023F9D0 A0A069DVA1 A0A2A3EMJ4 A0A088AKB0 A0A2P8YQ77 A0A1W4VH03 B4MGG1 B4K4E8 B5DSA5 B4GE72 A0A0M4ECL6 B3MTK8 B4JHQ2 B3NJ89 B4NV21 B4INN1 L7EEF9 Q8MQI6 D3TLC9 A0A1A9XEY4 A0A1B0ARE6 A0A1B0ADA5 A0A1A9W8D4 A0A1B0EZI1 A0A1B0CEK0 A0A0P4W1Q8 R4FMJ2 Q0IFZ5 A0A023EE27 A0A0L7R030 A0A1Q3F5H5 B0WEX0 A0A1B0FCQ6 B4NA07 A0A0J7NAY3 A0A026X1S1 A0A154PGE5 A0A195CT75 A0A1A9VQ55 T1H732 A0A0K8TP13 R4UJU5 A0A0L0CCD2 E9IRQ5 A0A0N0BF85 A0A151X763 T1JJN4 F4WZ99 A0A195F7Q9 A0A158P1I6 N6U4Q3 A0A1W4WPC6 A0A151IZ15 A0A182PTZ8 A0A154PL55 A0A2J7PHF4 A0A067QXF0 A0A1S3DTG5 J9KPW7 A0A087UK15 A0A1I8M637 A0A182YEV5 A0A182LZ49 A0A182S6S0 A0A182V886 A0A182UK62 A0A182KXY1 Q7PH52 A0A182R8D6 A0A182WVF1 E2AFU8 A0A1Y9HA44 A0A182W2Z8 A0A084WR31 A0A3S3SPR9 A0A1L8EDQ6 A0A2M3ZDH0 A0A2M4AYZ2 A0A2M4C506 W5JGJ2 A0A182FX91 A0A0L7QL26 A0A2R7WD39 E0VKM1 A0A293LKJ5
PDB
1WII
E-value=1.03773e-21,
Score=248
Ontologies
PANTHER
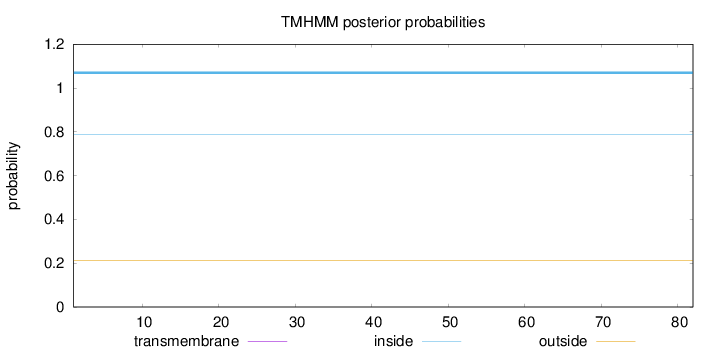
Topology
Subcellular location
Nucleus
Length:
82
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.78812
inside
1 - 82
Population Genetic Test Statistics
Pi
211.318223
Theta
173.48244
Tajima's D
0.583337
CLR
0
CSRT
0.539873006349682
Interpretation
Uncertain
