Gene
KWMTBOMO12963
Pre Gene Modal
BGIBMGA000448
Annotation
hypothetical_protein_RR46_08577_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 2.518
Sequence
CDS
ATGGCACATACCTTTTATTTAATATGTCTGCTTTGTATTTTTGGCGTAAGAATTACTACCGTTTCATCAGAATGTAATATTCCAATTGTATTGAGGAATACTTGGTTTTCTTTTGAGAATGGACGACAAACTGTAACTGATATTAATGCCAGCGATATGACCGGCAGAGGAGTGTGTGTGAATATCAAAGCGGAATTCCGGGTGAACTACACAATGGTGTTCCAATATACAAACTGTTATTATTGTGTTAAACTTATTGTACGAACTGTAAATGTTTTGGAGAAAATTGAAACTCCCTGTGTAGGGTTAGATCCAGATGAAGAACCAACTGTTGACAGAGTGTGTAGAGGCTTAAACCCTGATCAGAACCTAATTACGTTATTTTCTGAAAATTATGTGCCGGTCAACTGCCGTTCTTCTCTTGAGGGTGTATGGCAATTTGCATACCAAAACCGTTTTCGTTTCACTGGAGAATGCAATCATCCTGATGCCCAGATCAAATCCTGTCAAACTGCCGGAACTCAGTTTCTTATTACTAATCAAAAGTTTAATATAACATACAGGAGATGCCAAGGAATGTCTAATACTTTTGATGGTGTAGTTGAATACAGTTGTTTAGGTCATTGGTTTGTGGATAAGAATCACTTCTTTGCTGTCGCAAACACTAAGGAATCTCGTAAGGATGAGAAATACCGTTGTTTCCTCAAGAACAGAGATGATGACCTATACATTGGTGCATCCATCACTCCCGAGTGTAATACCCTGAAAACTGTGGAGAAATCACCCGAAAGATACAGAGTAACTCCTGTAAAGGCTGAAGTAGTAGAACCAGGTTGTCGCTTGCCACAGAACTTCTCTGGAGAATGGGTGAATACAGCCAATATTGATGCTGATGTATTTATTAATGAAACTCATATAATTGAAACATATTATCCCGATGAAGGCAGATACAGAAGAACTATTTATGTTTGCAGAGAACAACGGGATACTCGTGTAATGATGGCTCGTCTCACTGTTGATGGTTGCCAGAAGGATTACATATGTTTTGATTTCGTACCAAGGCATCACAATATCATACGGTATCGCAAAGGATTGGCCATGATTCAGAGCAACTTTCACACTGTGTGTTCCTGGGTACAGTTCCCGAATAAACAACAGTGGCGTTACGATATTTTTCTTAGGAAAGATCCATCACCTATTCGTTGCCCTGTAGCTGGAAAATTCAATTTTACCCAAAAAGGCGATGTAAAATTTGAAACCAGGATTCTCGGTGGAGTTACTTTGAGTCCTCGACCTAATTTGTATTGTAAAATCAATATAAGTGACTTCTCTGTGTGTGATTTTGATCAGAAAACTATACAAATAAAGGAAAATTATTGTCTGACAGTAGATTATTTGGGAAGACCAGTGGATATATACAGTGACCCAGATTACAAAATGAAATGTATTGGATATTGGAAGGAGAATTTGAAGTCATATTTGATAACATTTGATGAGTTAGATCCTTTCTCTAAATACAGGTGTTGGGTGTATCAAAGAGCTGATTTAAACAGAGTGTTAATGTCACAAGCCTTAGGTCCATACTGTGATTTAAAACAGGATGTCACATCATGGAATTACACAGAGGGGGCTGCTGTCGCCGTAGAAATGGAAGAATACGAGAGAGAAAGAGACCAGTGTCCTATGCATTTTGACGATGGCAGCGACCCTTGGAGTGCAAAAGAAAGCCTAATTCGAATTCTAACATACTCATATTGGAGTTCAAGCAGTGCCACTTTGCTAAGTATCAGTTTATCATCACTGATAATGGTGGCCTTAACTTTTGTAATACAATGA
Protein
MAHTFYLICLLCIFGVRITTVSSECNIPIVLRNTWFSFENGRQTVTDINASDMTGRGVCVNIKAEFRVNYTMVFQYTNCYYCVKLIVRTVNVLEKIETPCVGLDPDEEPTVDRVCRGLNPDQNLITLFSENYVPVNCRSSLEGVWQFAYQNRFRFTGECNHPDAQIKSCQTAGTQFLITNQKFNITYRRCQGMSNTFDGVVEYSCLGHWFVDKNHFFAVANTKESRKDEKYRCFLKNRDDDLYIGASITPECNTLKTVEKSPERYRVTPVKAEVVEPGCRLPQNFSGEWVNTANIDADVFINETHIIETYYPDEGRYRRTIYVCREQRDTRVMMARLTVDGCQKDYICFDFVPRHHNIIRYRKGLAMIQSNFHTVCSWVQFPNKQQWRYDIFLRKDPSPIRCPVAGKFNFTQKGDVKFETRILGGVTLSPRPNLYCKINISDFSVCDFDQKTIQIKENYCLTVDYLGRPVDIYSDPDYKMKCIGYWKENLKSYLITFDELDPFSKYRCWVYQRADLNRVLMSQALGPYCDLKQDVTSWNYTEGAAVAVEMEEYERERDQCPMHFDDGSDPWSAKESLIRILTYSYWSSSSATLLSISLSSLIMVALTFVIQ
Summary
Uniprot
A0A2H1W0G4
A0A2A4JA81
H9IT69
A0A194Q983
A0A212F4M8
A0A3S2TQM6
+ More
A0A194QXW2 A0A0L7LCZ6 A0A2J7QSD4 A0A067QJY8 A0A2A3ELC5 A0A088AIL4 A0A0L7RD64 A0A1W4XC47 A0A026X4K8 A0A1B6GH00 A0A1B6D081 A0A1B6J6L0 K7J7Y7 A0A1Y1LWZ2 A0A154PFX9 A0A232FGL7 E9J7U1 A0A2P8YJI9 A0A151IJT6 D6X3D9 F4WWB3 A0A1L8DP88 A0A195FBL6 A0A1L8DPA6 A0A158NKI5 A0A1B0CHS3 U4UFW7 N6T6K7 U5ESR6 A0A151WRN1 A0A146LII8 T1I229 A0A182XVD0 A0A0P6IXL4 A0A182NG79 N6U410 A0A0C9RD84 E2BW66 A0A2M4AF48 A0A182Q302 A0A182W2K8 A0A2M4BHI3 A0A182R7M0 A0A195B0P8 A0A182GH12 J9JKM8 W5JPM5 A0A182XLX2 A0A2M3ZE95 A0A2M3Z2V4 A0A182U477 A0A182IS79 A0A182FPY7 A0A182PEH5 A0A182LQB2 Q7QKE8 A0A182HS23 A0A182US06 A0A182K2W3 B0WXK7 A0A1Q3FGT5 B4K5B2 A0A1J1J0T9 Q9VCI4 A0A0P6FSN0 A0A1W4UKA6 B3LY51 A0A0P5Y2J7 A0A0P5S2N8 B4R246 E2AC68 B4M0F5 A0A0P5ZIB6 B4HFY4 B4JT96 A0A0P5SJN9 A0A1B0G2H5 A0A0P6FFS3 B3P8E3 A0A0P5PPN3 B4PNX6 A0A0P5AG45 A0A0P4ZE97 B4NG44 A0A0P5MED9 A0A0K8V4P1 A0A0A1WSL3 A0A182MM22 W8BZD6 A0A1I8NT13 A0A310S625 A0A1I8MCZ4 A0A1A9X5Z1 A0A2S2QBF7
A0A194QXW2 A0A0L7LCZ6 A0A2J7QSD4 A0A067QJY8 A0A2A3ELC5 A0A088AIL4 A0A0L7RD64 A0A1W4XC47 A0A026X4K8 A0A1B6GH00 A0A1B6D081 A0A1B6J6L0 K7J7Y7 A0A1Y1LWZ2 A0A154PFX9 A0A232FGL7 E9J7U1 A0A2P8YJI9 A0A151IJT6 D6X3D9 F4WWB3 A0A1L8DP88 A0A195FBL6 A0A1L8DPA6 A0A158NKI5 A0A1B0CHS3 U4UFW7 N6T6K7 U5ESR6 A0A151WRN1 A0A146LII8 T1I229 A0A182XVD0 A0A0P6IXL4 A0A182NG79 N6U410 A0A0C9RD84 E2BW66 A0A2M4AF48 A0A182Q302 A0A182W2K8 A0A2M4BHI3 A0A182R7M0 A0A195B0P8 A0A182GH12 J9JKM8 W5JPM5 A0A182XLX2 A0A2M3ZE95 A0A2M3Z2V4 A0A182U477 A0A182IS79 A0A182FPY7 A0A182PEH5 A0A182LQB2 Q7QKE8 A0A182HS23 A0A182US06 A0A182K2W3 B0WXK7 A0A1Q3FGT5 B4K5B2 A0A1J1J0T9 Q9VCI4 A0A0P6FSN0 A0A1W4UKA6 B3LY51 A0A0P5Y2J7 A0A0P5S2N8 B4R246 E2AC68 B4M0F5 A0A0P5ZIB6 B4HFY4 B4JT96 A0A0P5SJN9 A0A1B0G2H5 A0A0P6FFS3 B3P8E3 A0A0P5PPN3 B4PNX6 A0A0P5AG45 A0A0P4ZE97 B4NG44 A0A0P5MED9 A0A0K8V4P1 A0A0A1WSL3 A0A182MM22 W8BZD6 A0A1I8NT13 A0A310S625 A0A1I8MCZ4 A0A1A9X5Z1 A0A2S2QBF7
Pubmed
19121390
26354079
22118469
26227816
24845553
24508170
+ More
30249741 20075255 28004739 28648823 21282665 29403074 18362917 19820115 21719571 21347285 23537049 26823975 25244985 26999592 20798317 26483478 20920257 23761445 20966253 12364791 14747013 17210077 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25830018 24495485 25315136
30249741 20075255 28004739 28648823 21282665 29403074 18362917 19820115 21719571 21347285 23537049 26823975 25244985 26999592 20798317 26483478 20920257 23761445 20966253 12364791 14747013 17210077 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 25830018 24495485 25315136
EMBL
ODYU01005445
SOQ46312.1
NWSH01002266
PCG68749.1
BABH01013400
KQ459324
+ More
KPJ01540.1 AGBW02010334 OWR48698.1 RSAL01000021 RVE52480.1 KQ461073 KPJ09800.1 JTDY01001617 KOB73342.1 NEVH01011876 PNF31490.1 KK853376 KDR08043.1 KZ288222 PBC32072.1 KQ414614 KOC68907.1 KK107021 QOIP01000011 EZA62349.1 RLU16961.1 GECZ01015501 GECZ01008055 JAS54268.1 JAS61714.1 GEDC01018232 JAS19066.1 GECU01012900 JAS94806.1 GEZM01044893 JAV78059.1 KQ434896 KZC10769.1 NNAY01000231 OXU29835.1 GL768630 EFZ11112.1 PYGN01000551 PSN44406.1 KQ977294 KYN03910.1 KQ971372 EFA09764.1 GL888406 EGI61455.1 GFDF01005900 JAV08184.1 KQ981693 KYN37776.1 GFDF01005899 JAV08185.1 ADTU01018855 AJWK01012627 AJWK01012628 AJWK01012629 KB632336 ERL92844.1 APGK01050306 KB741169 ENN73323.1 GANO01004711 JAB55160.1 KQ982805 KYQ50468.1 GDHC01012349 JAQ06280.1 ACPB03000298 GDUN01000205 JAN95714.1 ENN73322.1 GBYB01006235 JAG76002.1 GL451091 EFN80098.1 GGFK01006083 MBW39404.1 AXCN02001629 GGFJ01003379 MBW52520.1 KQ976692 KYM77865.1 JXUM01062862 KQ562219 KXJ76367.1 ABLF02023612 ADMH02000471 ETN66342.1 GGFM01006105 MBW26856.1 GGFM01002095 MBW22846.1 AAAB01008799 EAA03780.3 APCN01000234 DS232168 EDS36537.1 GFDL01008274 JAV26771.1 CH933806 EDW16138.1 CVRI01000066 CRL05952.1 AE014297 BT033061 AAF56181.2 ACE82584.1 GDIQ01043365 JAN51372.1 CH902617 EDV43955.1 GDIP01064904 JAM38811.1 GDIQ01110489 GDIQ01106192 GDIP01045964 LRGB01002849 JAL41237.1 JAM57751.1 KZS06128.1 CM000364 EDX14103.1 GL438428 EFN68962.1 CH940650 EDW68334.1 GDIP01043240 JAM60475.1 CH480815 EDW43377.1 CH916373 EDV94986.1 GDIQ01094838 JAL56888.1 CCAG010018314 GDIQ01060511 JAN34226.1 CH954182 EDV53967.1 GDIQ01125221 JAL26505.1 CM000160 EDW98186.1 GDIP01199334 JAJ24068.1 GDIP01217930 JAJ05472.1 CH964251 EDW83261.2 GDIQ01157222 JAK94503.1 GDHF01018478 JAI33836.1 GBXI01012500 JAD01792.1 AXCM01000365 GAMC01004497 JAC02059.1 KQ768821 OAD53054.1 GGMS01005864 MBY75067.1
KPJ01540.1 AGBW02010334 OWR48698.1 RSAL01000021 RVE52480.1 KQ461073 KPJ09800.1 JTDY01001617 KOB73342.1 NEVH01011876 PNF31490.1 KK853376 KDR08043.1 KZ288222 PBC32072.1 KQ414614 KOC68907.1 KK107021 QOIP01000011 EZA62349.1 RLU16961.1 GECZ01015501 GECZ01008055 JAS54268.1 JAS61714.1 GEDC01018232 JAS19066.1 GECU01012900 JAS94806.1 GEZM01044893 JAV78059.1 KQ434896 KZC10769.1 NNAY01000231 OXU29835.1 GL768630 EFZ11112.1 PYGN01000551 PSN44406.1 KQ977294 KYN03910.1 KQ971372 EFA09764.1 GL888406 EGI61455.1 GFDF01005900 JAV08184.1 KQ981693 KYN37776.1 GFDF01005899 JAV08185.1 ADTU01018855 AJWK01012627 AJWK01012628 AJWK01012629 KB632336 ERL92844.1 APGK01050306 KB741169 ENN73323.1 GANO01004711 JAB55160.1 KQ982805 KYQ50468.1 GDHC01012349 JAQ06280.1 ACPB03000298 GDUN01000205 JAN95714.1 ENN73322.1 GBYB01006235 JAG76002.1 GL451091 EFN80098.1 GGFK01006083 MBW39404.1 AXCN02001629 GGFJ01003379 MBW52520.1 KQ976692 KYM77865.1 JXUM01062862 KQ562219 KXJ76367.1 ABLF02023612 ADMH02000471 ETN66342.1 GGFM01006105 MBW26856.1 GGFM01002095 MBW22846.1 AAAB01008799 EAA03780.3 APCN01000234 DS232168 EDS36537.1 GFDL01008274 JAV26771.1 CH933806 EDW16138.1 CVRI01000066 CRL05952.1 AE014297 BT033061 AAF56181.2 ACE82584.1 GDIQ01043365 JAN51372.1 CH902617 EDV43955.1 GDIP01064904 JAM38811.1 GDIQ01110489 GDIQ01106192 GDIP01045964 LRGB01002849 JAL41237.1 JAM57751.1 KZS06128.1 CM000364 EDX14103.1 GL438428 EFN68962.1 CH940650 EDW68334.1 GDIP01043240 JAM60475.1 CH480815 EDW43377.1 CH916373 EDV94986.1 GDIQ01094838 JAL56888.1 CCAG010018314 GDIQ01060511 JAN34226.1 CH954182 EDV53967.1 GDIQ01125221 JAL26505.1 CM000160 EDW98186.1 GDIP01199334 JAJ24068.1 GDIP01217930 JAJ05472.1 CH964251 EDW83261.2 GDIQ01157222 JAK94503.1 GDHF01018478 JAI33836.1 GBXI01012500 JAD01792.1 AXCM01000365 GAMC01004497 JAC02059.1 KQ768821 OAD53054.1 GGMS01005864 MBY75067.1
Proteomes
UP000218220
UP000005204
UP000053268
UP000007151
UP000283053
UP000053240
+ More
UP000037510 UP000235965 UP000027135 UP000242457 UP000005203 UP000053825 UP000192223 UP000053097 UP000279307 UP000002358 UP000076502 UP000215335 UP000245037 UP000078542 UP000007266 UP000007755 UP000078541 UP000005205 UP000092461 UP000030742 UP000019118 UP000075809 UP000015103 UP000076408 UP000075884 UP000008237 UP000075886 UP000075920 UP000075900 UP000078540 UP000069940 UP000249989 UP000007819 UP000000673 UP000076407 UP000075902 UP000075880 UP000069272 UP000075885 UP000075882 UP000007062 UP000075840 UP000075903 UP000075881 UP000002320 UP000009192 UP000183832 UP000000803 UP000192221 UP000007801 UP000076858 UP000000304 UP000000311 UP000008792 UP000001292 UP000001070 UP000092444 UP000008711 UP000002282 UP000007798 UP000075883 UP000095300 UP000095301 UP000092443
UP000037510 UP000235965 UP000027135 UP000242457 UP000005203 UP000053825 UP000192223 UP000053097 UP000279307 UP000002358 UP000076502 UP000215335 UP000245037 UP000078542 UP000007266 UP000007755 UP000078541 UP000005205 UP000092461 UP000030742 UP000019118 UP000075809 UP000015103 UP000076408 UP000075884 UP000008237 UP000075886 UP000075920 UP000075900 UP000078540 UP000069940 UP000249989 UP000007819 UP000000673 UP000076407 UP000075902 UP000075880 UP000069272 UP000075885 UP000075882 UP000007062 UP000075840 UP000075903 UP000075881 UP000002320 UP000009192 UP000183832 UP000000803 UP000192221 UP000007801 UP000076858 UP000000304 UP000000311 UP000008792 UP000001292 UP000001070 UP000092444 UP000008711 UP000002282 UP000007798 UP000075883 UP000095300 UP000095301 UP000092443
PRIDE
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2H1W0G4
A0A2A4JA81
H9IT69
A0A194Q983
A0A212F4M8
A0A3S2TQM6
+ More
A0A194QXW2 A0A0L7LCZ6 A0A2J7QSD4 A0A067QJY8 A0A2A3ELC5 A0A088AIL4 A0A0L7RD64 A0A1W4XC47 A0A026X4K8 A0A1B6GH00 A0A1B6D081 A0A1B6J6L0 K7J7Y7 A0A1Y1LWZ2 A0A154PFX9 A0A232FGL7 E9J7U1 A0A2P8YJI9 A0A151IJT6 D6X3D9 F4WWB3 A0A1L8DP88 A0A195FBL6 A0A1L8DPA6 A0A158NKI5 A0A1B0CHS3 U4UFW7 N6T6K7 U5ESR6 A0A151WRN1 A0A146LII8 T1I229 A0A182XVD0 A0A0P6IXL4 A0A182NG79 N6U410 A0A0C9RD84 E2BW66 A0A2M4AF48 A0A182Q302 A0A182W2K8 A0A2M4BHI3 A0A182R7M0 A0A195B0P8 A0A182GH12 J9JKM8 W5JPM5 A0A182XLX2 A0A2M3ZE95 A0A2M3Z2V4 A0A182U477 A0A182IS79 A0A182FPY7 A0A182PEH5 A0A182LQB2 Q7QKE8 A0A182HS23 A0A182US06 A0A182K2W3 B0WXK7 A0A1Q3FGT5 B4K5B2 A0A1J1J0T9 Q9VCI4 A0A0P6FSN0 A0A1W4UKA6 B3LY51 A0A0P5Y2J7 A0A0P5S2N8 B4R246 E2AC68 B4M0F5 A0A0P5ZIB6 B4HFY4 B4JT96 A0A0P5SJN9 A0A1B0G2H5 A0A0P6FFS3 B3P8E3 A0A0P5PPN3 B4PNX6 A0A0P5AG45 A0A0P4ZE97 B4NG44 A0A0P5MED9 A0A0K8V4P1 A0A0A1WSL3 A0A182MM22 W8BZD6 A0A1I8NT13 A0A310S625 A0A1I8MCZ4 A0A1A9X5Z1 A0A2S2QBF7
A0A194QXW2 A0A0L7LCZ6 A0A2J7QSD4 A0A067QJY8 A0A2A3ELC5 A0A088AIL4 A0A0L7RD64 A0A1W4XC47 A0A026X4K8 A0A1B6GH00 A0A1B6D081 A0A1B6J6L0 K7J7Y7 A0A1Y1LWZ2 A0A154PFX9 A0A232FGL7 E9J7U1 A0A2P8YJI9 A0A151IJT6 D6X3D9 F4WWB3 A0A1L8DP88 A0A195FBL6 A0A1L8DPA6 A0A158NKI5 A0A1B0CHS3 U4UFW7 N6T6K7 U5ESR6 A0A151WRN1 A0A146LII8 T1I229 A0A182XVD0 A0A0P6IXL4 A0A182NG79 N6U410 A0A0C9RD84 E2BW66 A0A2M4AF48 A0A182Q302 A0A182W2K8 A0A2M4BHI3 A0A182R7M0 A0A195B0P8 A0A182GH12 J9JKM8 W5JPM5 A0A182XLX2 A0A2M3ZE95 A0A2M3Z2V4 A0A182U477 A0A182IS79 A0A182FPY7 A0A182PEH5 A0A182LQB2 Q7QKE8 A0A182HS23 A0A182US06 A0A182K2W3 B0WXK7 A0A1Q3FGT5 B4K5B2 A0A1J1J0T9 Q9VCI4 A0A0P6FSN0 A0A1W4UKA6 B3LY51 A0A0P5Y2J7 A0A0P5S2N8 B4R246 E2AC68 B4M0F5 A0A0P5ZIB6 B4HFY4 B4JT96 A0A0P5SJN9 A0A1B0G2H5 A0A0P6FFS3 B3P8E3 A0A0P5PPN3 B4PNX6 A0A0P5AG45 A0A0P4ZE97 B4NG44 A0A0P5MED9 A0A0K8V4P1 A0A0A1WSL3 A0A182MM22 W8BZD6 A0A1I8NT13 A0A310S625 A0A1I8MCZ4 A0A1A9X5Z1 A0A2S2QBF7
Ontologies
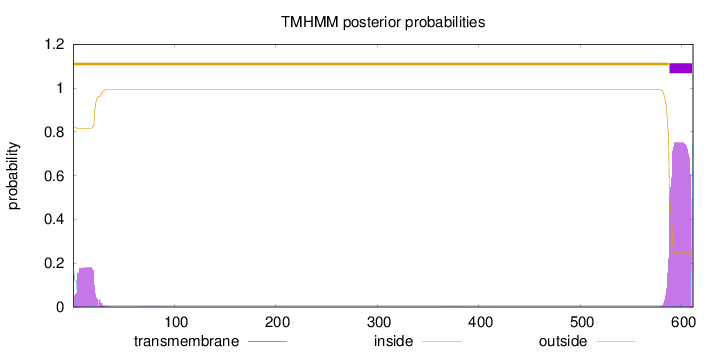
Topology
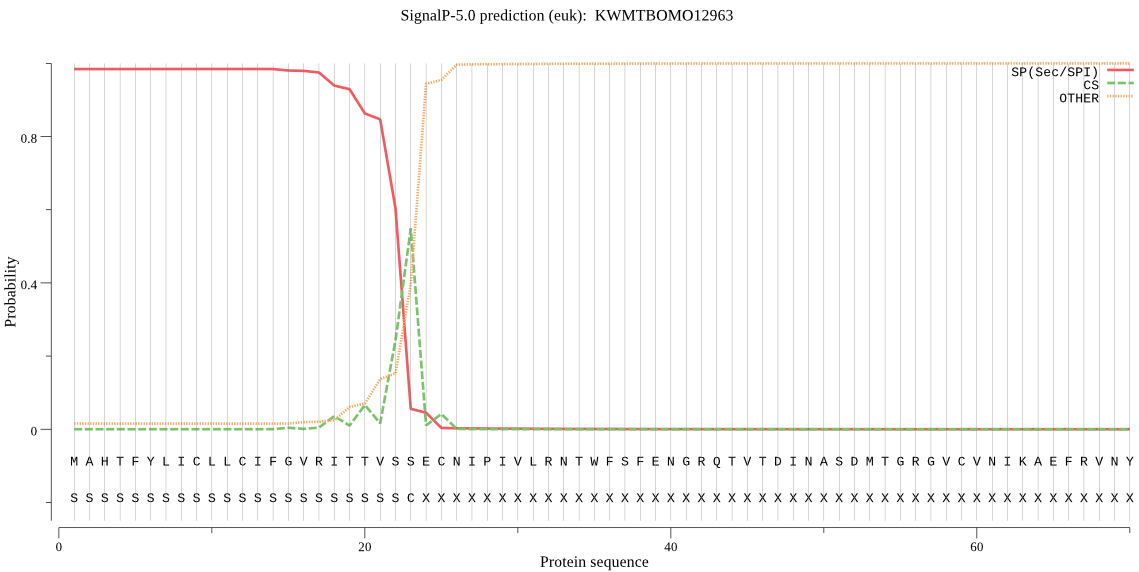
SignalP
Position: 1 - 23,
Likelihood: 0.984567
Length:
611
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.18296
Exp number, first 60 AAs:
3.53362
Total prob of N-in:
0.17985
outside
1 - 587
TMhelix
588 - 610
inside
611 - 611
Population Genetic Test Statistics
Pi
237.857008
Theta
196.676986
Tajima's D
0.639015
CLR
0.342296
CSRT
0.56147192640368
Interpretation
Uncertain
