Pre Gene Modal
BGIBMGA000452
Annotation
innexin_3_[Bombyx_mori]
Full name
Innexin
Location in the cell
PlasmaMembrane Reliability : 4.466
Sequence
CDS
ATGGCTGTTTTCGGTTTGGTATCATCAGTAGCGGGCTTCGTGAAAGTCCGCTATCTGATCGACAAGGCTGTCATCGATAACATGGTGTTCCGTATGCACTATCGGATCACATCGGCCATACTGTTTCTTTGTTGCATACTCGTTACGGCTAATAACCTGATCGGTGAACCCATTGCTTGCATTAGTGACGGCGCTAACCCTGGACACGTGATCAATACGTTCTGCTGGATCACGTACACATTCACCATGCCGAATACTACATCTAAAACCGCAGCCCATCCTGGTCTCGGCGACGACAATGATGAGAAACGCATACACTCTTACTATCAATGGGTACCGTTCATGCTGTTCTTCCAGGGCCTACTTTTTTACATTCCTCATTGGATTTGGAAGAACTGGGAAGAAGGAAAAGTCCGTCTGATTTCAGAAGGCATGAGAGGCACAATGGCCAGTATAGCTGATGACAAAAACAATCGTCAGAATCGACTCGTTCAATATTTGTTGGATACGTTGCACATGCACAACACTTACTCCTTCGGTTACTTCTTCTGCGAAGTGTTAAATTTCGCTAATGTCGTGGGAAACATATTTTTCTTGGACACATTCCTTGGCGGCGCATTCTTGACCTACGGCACCGACGTAGTCAGATTCTCCAATATGAATCAAGAGCAACGAACAGATCCAATGATCGAAGTTTTCCCGAGAATTACTAAATGTACATTCCATAAGTTCGGTGCCTCTGGAACGATACAAAAACACGACGCGCTTTGCGTCCTCGCTTTGAATATTCTTAACGAGAAAATTTTCATCTTCCTGTGGTTCTGGTTCATCATCCTGTCAGTGGTTTCCGGACTGGCACTGGTATACTCGGCTGCGGTATGTTTATTGCCAAGCACTCGTGAGACTATACTCAAAAGAAGATTCCGGTTCGGAACACCAAACGGCGTCGAAGCGCTCGTTAGGAAGACACAGGTCGGCGATTTCTTGCTTCTACATCTGCTGGGCCAGAATATGTCGTTGCGAGTGTTTGGTGAAGTTTTGGATGAGCTATCACGAAGACTTAATCTAGGTTCCCATGCTCCGTCAGCTCCGTCGACGCTCGAGATGGCTCCTATTTATCCTGACATTGACAAGTATTCTAAGGAAACCGAAACGTAA
Protein
MAVFGLVSSVAGFVKVRYLIDKAVIDNMVFRMHYRITSAILFLCCILVTANNLIGEPIACISDGANPGHVINTFCWITYTFTMPNTTSKTAAHPGLGDDNDEKRIHSYYQWVPFMLFFQGLLFYIPHWIWKNWEEGKVRLISEGMRGTMASIADDKNNRQNRLVQYLLDTLHMHNTYSFGYFFCEVLNFANVVGNIFFLDTFLGGAFLTYGTDVVRFSNMNQEQRTDPMIEVFPRITKCTFHKFGASGTIQKHDALCVLALNILNEKIFIFLWFWFIILSVVSGLALVYSAAVCLLPSTRETILKRRFRFGTPNGVEALVRKTQVGDFLLLHLLGQNMSLRVFGEVLDELSRRLNLGSHAPSAPSTLEMAPIYPDIDKYSKETET
Summary
Description
Structural component of the gap junctions.
Similarity
Belongs to the pannexin family.
Feature
chain Innexin
Uniprot
H9IT73
B7U9X6
A0A2A4K0X0
K9RZK7
S4PBP2
A0A3G1PLL2
+ More
A0A1E1WI21 I4DN96 A0A194QW93 A0A2H1VX70 A0A212F4N9 A0A194Q988 A0A1W4XSA6 D6X0D3 A0A232FC75 E2AUZ7 A0A088AID2 A0A2A3EKM6 E2BAV2 A0A154P9Y4 A0A151X8N2 A0A0C9QJQ2 A0A310SDX5 A0A0J7KFC1 A0A195ATV3 A0A158NPV6 A0A151IHI3 F4W7D6 A0A0T6AWJ1 A0A151JSR9 A0A026WWJ5 A0A195DTQ6 A0A2P8YHY4 A0A1B6KPH5 A0A1B6GUF3 A0A182PP64 A0A182NGH0 A0A182M1Z1 A0A182JRU8 A0A182Y8R0 A0A2M3Z359 A0A2M4BQU8 A0A182RE29 A0A182W3H4 A0A2M4A5M4 A0A182FAM7 W5J1S6 A0A182TQV8 A0A023ESH3 A0A182QG05 A0A182HF50 A0A182L2S3 A0A182HYQ5 Q16QK9 A0A182XIH4 T1E2Q0 A0A075M4U6 A0A1S4FT50 A0A0L7RDI7 A0A182V5Z4 E0VNL8 Q7Q9Z6 A0A075M4I1 A0A1Q3FNY4 A0A1B6BWP0 A0A1Q3FP12 A0A2J7PSE5 B0X6R5 T1D6D2 U5EZ77 A0A182JA93 A0A1S3CXD3 A0A023F482 A0A1L8E367 T1I6M9 A0A0V0G6T3 Q5XUA2 A0A1B6JAV4 A0A2S2QIB7 A0A2H8TSZ7 A0A224XDV8 A0A2S2NNK0 J9JWH0 A0A069DTP9 A0A336M2F4 A0A0A9WAN6 A0A1B0CI13 N6U8L4 U4UDN4 A0A1J1JAC4 A0A1I8MBQ8 A0A1I8P752 A0A1A9WEF9 A0A1B0FDH7 A0A1A9VAH4 A0A1B0A0G2 A0A182SCY0 A0A1Q3FNX7 B4NKS4 A0A0M9A171
A0A1E1WI21 I4DN96 A0A194QW93 A0A2H1VX70 A0A212F4N9 A0A194Q988 A0A1W4XSA6 D6X0D3 A0A232FC75 E2AUZ7 A0A088AID2 A0A2A3EKM6 E2BAV2 A0A154P9Y4 A0A151X8N2 A0A0C9QJQ2 A0A310SDX5 A0A0J7KFC1 A0A195ATV3 A0A158NPV6 A0A151IHI3 F4W7D6 A0A0T6AWJ1 A0A151JSR9 A0A026WWJ5 A0A195DTQ6 A0A2P8YHY4 A0A1B6KPH5 A0A1B6GUF3 A0A182PP64 A0A182NGH0 A0A182M1Z1 A0A182JRU8 A0A182Y8R0 A0A2M3Z359 A0A2M4BQU8 A0A182RE29 A0A182W3H4 A0A2M4A5M4 A0A182FAM7 W5J1S6 A0A182TQV8 A0A023ESH3 A0A182QG05 A0A182HF50 A0A182L2S3 A0A182HYQ5 Q16QK9 A0A182XIH4 T1E2Q0 A0A075M4U6 A0A1S4FT50 A0A0L7RDI7 A0A182V5Z4 E0VNL8 Q7Q9Z6 A0A075M4I1 A0A1Q3FNY4 A0A1B6BWP0 A0A1Q3FP12 A0A2J7PSE5 B0X6R5 T1D6D2 U5EZ77 A0A182JA93 A0A1S3CXD3 A0A023F482 A0A1L8E367 T1I6M9 A0A0V0G6T3 Q5XUA2 A0A1B6JAV4 A0A2S2QIB7 A0A2H8TSZ7 A0A224XDV8 A0A2S2NNK0 J9JWH0 A0A069DTP9 A0A336M2F4 A0A0A9WAN6 A0A1B0CI13 N6U8L4 U4UDN4 A0A1J1JAC4 A0A1I8MBQ8 A0A1I8P752 A0A1A9WEF9 A0A1B0FDH7 A0A1A9VAH4 A0A1B0A0G2 A0A182SCY0 A0A1Q3FNX7 B4NKS4 A0A0M9A171
Pubmed
19121390
19415533
23622113
29361596
22651552
26354079
+ More
22118469 18362917 19820115 28648823 20798317 21347285 21719571 24508170 30249741 29403074 25244985 20920257 23761445 24945155 26483478 20966253 17510324 24330624 25585357 20566863 12364791 25474469 26334808 25401762 26823975 23537049 25315136 17994087
22118469 18362917 19820115 28648823 20798317 21347285 21719571 24508170 30249741 29403074 25244985 20920257 23761445 24945155 26483478 20966253 17510324 24330624 25585357 20566863 12364791 25474469 26334808 25401762 26823975 23537049 25315136 17994087
EMBL
BABH01013423
BABH01013424
FJ463039
ACK38254.1
NWSH01000333
PCG77320.1
+ More
KC018472 AFY62976.1 GAIX01004521 JAA88039.1 MG000944 AVM81262.1 GDQN01004415 JAT86639.1 AK402784 BAM19386.1 KQ461073 KPJ09808.1 ODYU01005002 SOQ45447.1 AGBW02010334 OWR48705.1 KQ459324 KPJ01550.1 KQ971372 EFA09591.1 NNAY01000509 OXU27937.1 GL442958 EFN62743.1 KZ288222 PBC32044.1 GL446860 EFN87180.1 KQ434856 KZC08746.1 KQ982409 KYQ56736.1 GBYB01003744 JAG73511.1 KQ760869 OAD58782.1 LBMM01008141 KMQ89113.1 KQ976745 KYM75487.1 ADTU01022766 KQ977606 KYN01423.1 GL887844 EGI69809.1 LJIG01022631 KRT79535.1 KQ982014 KYN30623.1 KK107105 QOIP01000011 EZA59499.1 RLU17101.1 KQ980487 KYN15909.1 PYGN01000582 PSN43853.1 GEBQ01026641 GEBQ01025246 GEBQ01023088 JAT13336.1 JAT14731.1 JAT16889.1 GECZ01003701 JAS66068.1 AXCM01004448 GGFM01002184 MBW22935.1 GGFJ01006251 MBW55392.1 GGFK01002607 MBW35928.1 ADMH02002159 ETN58197.1 GAPW01001882 JAC11716.1 AXCN02001607 JXUM01133535 KQ567992 KXJ69206.1 APCN01005396 CH477745 EAT36684.1 GALA01001095 JAA93757.1 KJ736825 AIF75097.1 KQ414614 KOC68874.1 AAZO01003917 DS235340 EEB14974.1 AAAB01008898 EAA09228.3 KJ736826 AIF75098.1 GFDL01005768 JAV29277.1 GEDC01031602 JAS05696.1 GFDL01005720 JAV29325.1 NEVH01021931 PNF19253.1 DS232421 EDS41548.1 GALA01000113 JAA94739.1 GANO01001289 JAB58582.1 GBBI01002387 GEMB01001828 JAC16325.1 JAS01343.1 GFDF01000923 JAV13161.1 ACPB03000870 GECL01002297 JAP03827.1 AY737552 AAU84945.1 GECU01037337 GECU01035501 GECU01011461 JAS70369.1 JAS72205.1 JAS96245.1 GGMS01008265 MBY77468.1 GFXV01002065 GFXV01005548 MBW13870.1 MBW17353.1 GFTR01005770 JAW10656.1 GGMR01006150 MBY18769.1 ABLF02034090 GBGD01001748 JAC87141.1 UFQS01000109 UFQT01000109 SSW99768.1 SSX20148.1 GBHO01040051 GDHC01010917 JAG03553.1 JAQ07712.1 AJWK01012959 APGK01044510 APGK01044511 KB741026 ENN74942.1 KB632309 ERL92084.1 CVRI01000075 CRL08502.1 CCAG010013497 GFDL01005750 JAV29295.1 CH964272 EDW84135.1 KQ435774 KOX75087.1
KC018472 AFY62976.1 GAIX01004521 JAA88039.1 MG000944 AVM81262.1 GDQN01004415 JAT86639.1 AK402784 BAM19386.1 KQ461073 KPJ09808.1 ODYU01005002 SOQ45447.1 AGBW02010334 OWR48705.1 KQ459324 KPJ01550.1 KQ971372 EFA09591.1 NNAY01000509 OXU27937.1 GL442958 EFN62743.1 KZ288222 PBC32044.1 GL446860 EFN87180.1 KQ434856 KZC08746.1 KQ982409 KYQ56736.1 GBYB01003744 JAG73511.1 KQ760869 OAD58782.1 LBMM01008141 KMQ89113.1 KQ976745 KYM75487.1 ADTU01022766 KQ977606 KYN01423.1 GL887844 EGI69809.1 LJIG01022631 KRT79535.1 KQ982014 KYN30623.1 KK107105 QOIP01000011 EZA59499.1 RLU17101.1 KQ980487 KYN15909.1 PYGN01000582 PSN43853.1 GEBQ01026641 GEBQ01025246 GEBQ01023088 JAT13336.1 JAT14731.1 JAT16889.1 GECZ01003701 JAS66068.1 AXCM01004448 GGFM01002184 MBW22935.1 GGFJ01006251 MBW55392.1 GGFK01002607 MBW35928.1 ADMH02002159 ETN58197.1 GAPW01001882 JAC11716.1 AXCN02001607 JXUM01133535 KQ567992 KXJ69206.1 APCN01005396 CH477745 EAT36684.1 GALA01001095 JAA93757.1 KJ736825 AIF75097.1 KQ414614 KOC68874.1 AAZO01003917 DS235340 EEB14974.1 AAAB01008898 EAA09228.3 KJ736826 AIF75098.1 GFDL01005768 JAV29277.1 GEDC01031602 JAS05696.1 GFDL01005720 JAV29325.1 NEVH01021931 PNF19253.1 DS232421 EDS41548.1 GALA01000113 JAA94739.1 GANO01001289 JAB58582.1 GBBI01002387 GEMB01001828 JAC16325.1 JAS01343.1 GFDF01000923 JAV13161.1 ACPB03000870 GECL01002297 JAP03827.1 AY737552 AAU84945.1 GECU01037337 GECU01035501 GECU01011461 JAS70369.1 JAS72205.1 JAS96245.1 GGMS01008265 MBY77468.1 GFXV01002065 GFXV01005548 MBW13870.1 MBW17353.1 GFTR01005770 JAW10656.1 GGMR01006150 MBY18769.1 ABLF02034090 GBGD01001748 JAC87141.1 UFQS01000109 UFQT01000109 SSW99768.1 SSX20148.1 GBHO01040051 GDHC01010917 JAG03553.1 JAQ07712.1 AJWK01012959 APGK01044510 APGK01044511 KB741026 ENN74942.1 KB632309 ERL92084.1 CVRI01000075 CRL08502.1 CCAG010013497 GFDL01005750 JAV29295.1 CH964272 EDW84135.1 KQ435774 KOX75087.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000192223
+ More
UP000007266 UP000215335 UP000000311 UP000005203 UP000242457 UP000008237 UP000076502 UP000075809 UP000036403 UP000078540 UP000005205 UP000078542 UP000007755 UP000078541 UP000053097 UP000279307 UP000078492 UP000245037 UP000075885 UP000075884 UP000075883 UP000075881 UP000076408 UP000075900 UP000075920 UP000069272 UP000000673 UP000075902 UP000075886 UP000069940 UP000249989 UP000075882 UP000075840 UP000008820 UP000076407 UP000053825 UP000075903 UP000009046 UP000007062 UP000235965 UP000002320 UP000075880 UP000079169 UP000015103 UP000007819 UP000092461 UP000019118 UP000030742 UP000183832 UP000095301 UP000095300 UP000091820 UP000092444 UP000078200 UP000092445 UP000075901 UP000007798 UP000053105
UP000007266 UP000215335 UP000000311 UP000005203 UP000242457 UP000008237 UP000076502 UP000075809 UP000036403 UP000078540 UP000005205 UP000078542 UP000007755 UP000078541 UP000053097 UP000279307 UP000078492 UP000245037 UP000075885 UP000075884 UP000075883 UP000075881 UP000076408 UP000075900 UP000075920 UP000069272 UP000000673 UP000075902 UP000075886 UP000069940 UP000249989 UP000075882 UP000075840 UP000008820 UP000076407 UP000053825 UP000075903 UP000009046 UP000007062 UP000235965 UP000002320 UP000075880 UP000079169 UP000015103 UP000007819 UP000092461 UP000019118 UP000030742 UP000183832 UP000095301 UP000095300 UP000091820 UP000092444 UP000078200 UP000092445 UP000075901 UP000007798 UP000053105
Pfam
PF00876 Innexin
Interpro
IPR000990
Innexin
ProteinModelPortal
H9IT73
B7U9X6
A0A2A4K0X0
K9RZK7
S4PBP2
A0A3G1PLL2
+ More
A0A1E1WI21 I4DN96 A0A194QW93 A0A2H1VX70 A0A212F4N9 A0A194Q988 A0A1W4XSA6 D6X0D3 A0A232FC75 E2AUZ7 A0A088AID2 A0A2A3EKM6 E2BAV2 A0A154P9Y4 A0A151X8N2 A0A0C9QJQ2 A0A310SDX5 A0A0J7KFC1 A0A195ATV3 A0A158NPV6 A0A151IHI3 F4W7D6 A0A0T6AWJ1 A0A151JSR9 A0A026WWJ5 A0A195DTQ6 A0A2P8YHY4 A0A1B6KPH5 A0A1B6GUF3 A0A182PP64 A0A182NGH0 A0A182M1Z1 A0A182JRU8 A0A182Y8R0 A0A2M3Z359 A0A2M4BQU8 A0A182RE29 A0A182W3H4 A0A2M4A5M4 A0A182FAM7 W5J1S6 A0A182TQV8 A0A023ESH3 A0A182QG05 A0A182HF50 A0A182L2S3 A0A182HYQ5 Q16QK9 A0A182XIH4 T1E2Q0 A0A075M4U6 A0A1S4FT50 A0A0L7RDI7 A0A182V5Z4 E0VNL8 Q7Q9Z6 A0A075M4I1 A0A1Q3FNY4 A0A1B6BWP0 A0A1Q3FP12 A0A2J7PSE5 B0X6R5 T1D6D2 U5EZ77 A0A182JA93 A0A1S3CXD3 A0A023F482 A0A1L8E367 T1I6M9 A0A0V0G6T3 Q5XUA2 A0A1B6JAV4 A0A2S2QIB7 A0A2H8TSZ7 A0A224XDV8 A0A2S2NNK0 J9JWH0 A0A069DTP9 A0A336M2F4 A0A0A9WAN6 A0A1B0CI13 N6U8L4 U4UDN4 A0A1J1JAC4 A0A1I8MBQ8 A0A1I8P752 A0A1A9WEF9 A0A1B0FDH7 A0A1A9VAH4 A0A1B0A0G2 A0A182SCY0 A0A1Q3FNX7 B4NKS4 A0A0M9A171
A0A1E1WI21 I4DN96 A0A194QW93 A0A2H1VX70 A0A212F4N9 A0A194Q988 A0A1W4XSA6 D6X0D3 A0A232FC75 E2AUZ7 A0A088AID2 A0A2A3EKM6 E2BAV2 A0A154P9Y4 A0A151X8N2 A0A0C9QJQ2 A0A310SDX5 A0A0J7KFC1 A0A195ATV3 A0A158NPV6 A0A151IHI3 F4W7D6 A0A0T6AWJ1 A0A151JSR9 A0A026WWJ5 A0A195DTQ6 A0A2P8YHY4 A0A1B6KPH5 A0A1B6GUF3 A0A182PP64 A0A182NGH0 A0A182M1Z1 A0A182JRU8 A0A182Y8R0 A0A2M3Z359 A0A2M4BQU8 A0A182RE29 A0A182W3H4 A0A2M4A5M4 A0A182FAM7 W5J1S6 A0A182TQV8 A0A023ESH3 A0A182QG05 A0A182HF50 A0A182L2S3 A0A182HYQ5 Q16QK9 A0A182XIH4 T1E2Q0 A0A075M4U6 A0A1S4FT50 A0A0L7RDI7 A0A182V5Z4 E0VNL8 Q7Q9Z6 A0A075M4I1 A0A1Q3FNY4 A0A1B6BWP0 A0A1Q3FP12 A0A2J7PSE5 B0X6R5 T1D6D2 U5EZ77 A0A182JA93 A0A1S3CXD3 A0A023F482 A0A1L8E367 T1I6M9 A0A0V0G6T3 Q5XUA2 A0A1B6JAV4 A0A2S2QIB7 A0A2H8TSZ7 A0A224XDV8 A0A2S2NNK0 J9JWH0 A0A069DTP9 A0A336M2F4 A0A0A9WAN6 A0A1B0CI13 N6U8L4 U4UDN4 A0A1J1JAC4 A0A1I8MBQ8 A0A1I8P752 A0A1A9WEF9 A0A1B0FDH7 A0A1A9VAH4 A0A1B0A0G2 A0A182SCY0 A0A1Q3FNX7 B4NKS4 A0A0M9A171
PDB
5H1R
E-value=6.9013e-12,
Score=170
Ontologies
GO
PANTHER
Topology
Subcellular location
Cell membrane
Cell junction
Gap junction
Cell junction
Gap junction
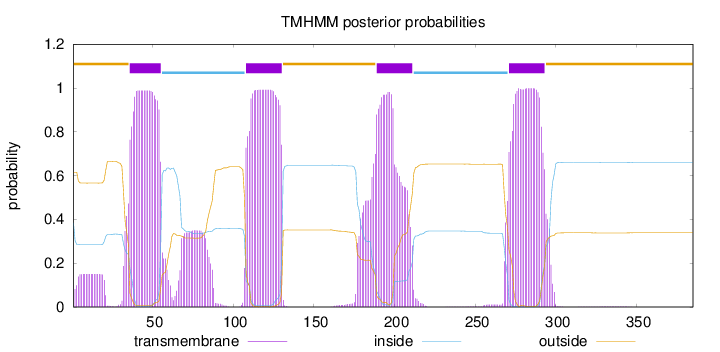
Length:
385
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
99.9058
Exp number, first 60 AAs:
23.94266
Total prob of N-in:
0.38424
POSSIBLE N-term signal
sequence
outside
1 - 35
TMhelix
36 - 55
inside
56 - 107
TMhelix
108 - 130
outside
131 - 188
TMhelix
189 - 211
inside
212 - 270
TMhelix
271 - 293
outside
294 - 385
Population Genetic Test Statistics
Pi
164.820777
Theta
173.232782
Tajima's D
-0.378298
CLR
1.135594
CSRT
0.271236438178091
Interpretation
Uncertain
