Gene
KWMTBOMO12958
Pre Gene Modal
BGIBMGA000240
Annotation
PREDICTED:_dedicator_of_cytokinesis_protein_3-like_[Papilio_polytes]
Location in the cell
Mitochondrial Reliability : 1.906
Sequence
CDS
ATGTGGATACCATCGAAAACTCCGAAACTAGCCGTCGCGGTCTACAACTGGAAAGGGGATGTTCGATCAGGTTTACCTTTGGAAATAGGTGAAACTGTACAGATATTGGAGGAAAATGGAGCTTGGTTCCGTGGGTTCAGCACAAAAAACAGATCAACATGGGGTATTTTTCCTGCCTGTGTTGTGTCTCTCAGACCGTGCACTGTGAAAGGCTCTGGGGCAACAGCTGTTGCTGAATTGAAGGATGATCCTCTAGTTCGTGAAATTGCATGTGTATTGAGAGAATGGGCTAGGCTTTGGAAGAAATTATATGTGGAACGTGAAACGTACAGATTCAGCGCAGTTGCGAAGGTAATGAGGGAACTGTTAAGTGGACGGCGTGCTCTTCTCGCGGGGACCCTAACACAAGATCAGACTAGGGCGCTAAGACTGAAGCTTGTTTCTAAACTAGATTGGGGCAATAGGTAA
Protein
MWIPSKTPKLAVAVYNWKGDVRSGLPLEIGETVQILEENGAWFRGFSTKNRSTWGIFPACVVSLRPCTVKGSGATAVAELKDDPLVREIACVLREWARLWKKLYVERETYRFSAVAKVMRELLSGRRALLAGTLTQDQTRALRLKLVSKLDWGNR
Summary
Similarity
Belongs to the DOCK family.
Uniprot
A0A194Q830
A0A2A4JZT5
A0A3S2PIN2
A0A194QWX7
A0A139WBU9
F4W7D7
+ More
A0A195ATG9 A0A0C9RFH9 A0A0C9QTT3 V5H0L9 A0A088AIJ0 A0A151X8N0 A0A158NPV7 A0A151JT19 A0A1Y1KUL2 A0A1Y1KUK7 A0A3L8D806 A0A195DSL5 A0A1Y1KX53 A0A1B6BYQ6 A0A1B6CEC0 A0A1Y1KUM5 A0A1W4XHM2 A0A1B6J4N1 A0A1B6ILA6 A0A310SNE4 A0A1B6LK86 A0A2H8TWY4 A0A2S2PUB0 A0A0N0BGP5 A0A232FBC8 A0A2H8TGS0 A0A2S2R2L6 A0A067R3F9 A0A2A3EJQ7 A0A026WU37 A0A0L7RDF3 K7J6U9 A0A023F3N0 A0A0A9Y114 T1I6Z7 A0A146L604 A0A3Q0INC7 A0A0T6AWV2 E0VNL7 A0A1B6GXX3 A0A0L0BYF7 A0A0M4ENT6 A0A151IHE9 A0A1Q3G2A9 A0A224XV80 A0A1B0DHS1 A0A0S7LZL0 A0A1I8MCC3 A0A1I8MCB9 A0A336LU26 A0A182HYQ6 A0A182W3H5 A0A1S4G791 B4M167 B3M338 A0A1A8G3P1 A0A1J1HPL1 B4NKS3 A0A1A8CPG8 A0A1A8JPJ1 A0A1Q3F7G8 A0A1Q3F7M9 A0A1S3K6I7 B4K955 A0A182GV94 A0A2I4BUX1 A0A1A9WEG1 A0A1W4VU94 A0A1W4VTJ0 A0A182PP63 A0A1A9VAI0 A0A1A8HBV3 A0A1A8GYZ2 W8BGP2 A0A1A8UI27 A0A182HF49 A0A1B0A0G7 A0A182Y8Q9 Q298R8 I3J9T5 A0A1B0BML3 Q16QL0 B7Z0R2 A0A0B4KHD2 Q9VAS8 A0A0K8VZL8 A0A182FAM6 A0A3Q3GU39 A0A3Q3BAZ6 A0A182JRU7 A0A3B5RCJ7 A0A3B5Q0N2
A0A195ATG9 A0A0C9RFH9 A0A0C9QTT3 V5H0L9 A0A088AIJ0 A0A151X8N0 A0A158NPV7 A0A151JT19 A0A1Y1KUL2 A0A1Y1KUK7 A0A3L8D806 A0A195DSL5 A0A1Y1KX53 A0A1B6BYQ6 A0A1B6CEC0 A0A1Y1KUM5 A0A1W4XHM2 A0A1B6J4N1 A0A1B6ILA6 A0A310SNE4 A0A1B6LK86 A0A2H8TWY4 A0A2S2PUB0 A0A0N0BGP5 A0A232FBC8 A0A2H8TGS0 A0A2S2R2L6 A0A067R3F9 A0A2A3EJQ7 A0A026WU37 A0A0L7RDF3 K7J6U9 A0A023F3N0 A0A0A9Y114 T1I6Z7 A0A146L604 A0A3Q0INC7 A0A0T6AWV2 E0VNL7 A0A1B6GXX3 A0A0L0BYF7 A0A0M4ENT6 A0A151IHE9 A0A1Q3G2A9 A0A224XV80 A0A1B0DHS1 A0A0S7LZL0 A0A1I8MCC3 A0A1I8MCB9 A0A336LU26 A0A182HYQ6 A0A182W3H5 A0A1S4G791 B4M167 B3M338 A0A1A8G3P1 A0A1J1HPL1 B4NKS3 A0A1A8CPG8 A0A1A8JPJ1 A0A1Q3F7G8 A0A1Q3F7M9 A0A1S3K6I7 B4K955 A0A182GV94 A0A2I4BUX1 A0A1A9WEG1 A0A1W4VU94 A0A1W4VTJ0 A0A182PP63 A0A1A9VAI0 A0A1A8HBV3 A0A1A8GYZ2 W8BGP2 A0A1A8UI27 A0A182HF49 A0A1B0A0G7 A0A182Y8Q9 Q298R8 I3J9T5 A0A1B0BML3 Q16QL0 B7Z0R2 A0A0B4KHD2 Q9VAS8 A0A0K8VZL8 A0A182FAM6 A0A3Q3GU39 A0A3Q3BAZ6 A0A182JRU7 A0A3B5RCJ7 A0A3B5Q0N2
Pubmed
26354079
18362917
19820115
21719571
21347285
28004739
+ More
30249741 28648823 24845553 24508170 20075255 25474469 25401762 26823975 20566863 26108605 25315136 17510324 17994087 26483478 24495485 25244985 15632085 25186727 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23542700
30249741 28648823 24845553 24508170 20075255 25474469 25401762 26823975 20566863 26108605 25315136 17510324 17994087 26483478 24495485 25244985 15632085 25186727 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 23542700
EMBL
KQ459324
KPJ01549.1
NWSH01000333
PCG77319.1
RSAL01000021
RVE52484.1
+ More
KQ461073 KPJ09809.1 KQ971372 KYB25392.1 GL887844 EGI69810.1 KQ976745 KYM75486.1 GBYB01007150 JAG76917.1 GBYB01007149 JAG76916.1 GALX01000669 JAB67797.1 KQ982409 KYQ56735.1 ADTU01022763 ADTU01022764 ADTU01022765 ADTU01022766 ADTU01022767 ADTU01022768 KQ982014 KYN30621.1 GEZM01073449 JAV65089.1 GEZM01073452 JAV65083.1 QOIP01000011 RLU16401.1 KQ980487 KYN15910.1 GEZM01073454 JAV65081.1 GEDC01030891 JAS06407.1 GEDC01025543 GEDC01015790 JAS11755.1 JAS21508.1 GEZM01073453 JAV65082.1 GECU01013589 JAS94117.1 GECU01019996 JAS87710.1 KQ760869 OAD58781.1 GEBQ01015887 JAT24090.1 GFXV01006941 MBW18746.1 GGMR01020346 MBY32965.1 KQ435774 KOX75088.1 NNAY01000509 OXU27932.1 GFXV01001434 MBW13239.1 GGMS01015078 MBY84281.1 KK852782 KDR16623.1 KZ288222 PBC32043.1 KK107105 EZA59498.1 KQ414614 KOC68873.1 AAZX01001292 AAZX01006521 AAZX01006718 AAZX01006766 AAZX01013753 AAZX01024928 GBBI01002727 JAC15985.1 GBHO01020424 JAG23180.1 ACPB03000870 GDHC01015973 JAQ02656.1 LJIG01022631 KRT79537.1 DS235340 EEB14973.1 GECZ01002496 JAS67273.1 JRES01001160 KNC25073.1 CP012526 ALC45635.1 KQ977606 KYN01422.1 GFDL01001105 JAV33940.1 GFTR01002688 JAW13738.1 AJVK01061505 GBYX01135134 JAO94787.1 UFQT01000109 SSX20149.1 APCN01005397 APCN01005398 APCN01005399 CH940650 EDW67478.2 CH902617 EDV42438.2 HAEB01019773 SBQ66300.1 CVRI01000006 CRK88153.1 CH964272 EDW84134.2 HADZ01016734 SBP80675.1 HAEE01001817 SBR21837.1 GFDL01011531 JAV23514.1 GFDL01011516 JAV23529.1 CH933806 EDW14468.2 JXUM01090538 KQ563846 KXJ73176.1 HAEC01013444 SBQ81661.1 HAEC01008141 SBQ76279.1 GAMC01010417 JAB96138.1 HAEJ01007273 SBS47730.1 JXUM01133534 KQ567992 KXJ69205.1 CM000070 EAL27887.4 AERX01048904 AERX01048905 AERX01048906 AERX01048907 AERX01048908 AERX01048909 AERX01048910 AERX01048911 AERX01048912 AERX01048913 JXJN01016991 CH477745 EAT36683.1 AE014297 ACL83574.2 AGB96429.1 AGB96427.1 AGB96428.1 AAF56821.5 GDHF01008033 JAI44281.1
KQ461073 KPJ09809.1 KQ971372 KYB25392.1 GL887844 EGI69810.1 KQ976745 KYM75486.1 GBYB01007150 JAG76917.1 GBYB01007149 JAG76916.1 GALX01000669 JAB67797.1 KQ982409 KYQ56735.1 ADTU01022763 ADTU01022764 ADTU01022765 ADTU01022766 ADTU01022767 ADTU01022768 KQ982014 KYN30621.1 GEZM01073449 JAV65089.1 GEZM01073452 JAV65083.1 QOIP01000011 RLU16401.1 KQ980487 KYN15910.1 GEZM01073454 JAV65081.1 GEDC01030891 JAS06407.1 GEDC01025543 GEDC01015790 JAS11755.1 JAS21508.1 GEZM01073453 JAV65082.1 GECU01013589 JAS94117.1 GECU01019996 JAS87710.1 KQ760869 OAD58781.1 GEBQ01015887 JAT24090.1 GFXV01006941 MBW18746.1 GGMR01020346 MBY32965.1 KQ435774 KOX75088.1 NNAY01000509 OXU27932.1 GFXV01001434 MBW13239.1 GGMS01015078 MBY84281.1 KK852782 KDR16623.1 KZ288222 PBC32043.1 KK107105 EZA59498.1 KQ414614 KOC68873.1 AAZX01001292 AAZX01006521 AAZX01006718 AAZX01006766 AAZX01013753 AAZX01024928 GBBI01002727 JAC15985.1 GBHO01020424 JAG23180.1 ACPB03000870 GDHC01015973 JAQ02656.1 LJIG01022631 KRT79537.1 DS235340 EEB14973.1 GECZ01002496 JAS67273.1 JRES01001160 KNC25073.1 CP012526 ALC45635.1 KQ977606 KYN01422.1 GFDL01001105 JAV33940.1 GFTR01002688 JAW13738.1 AJVK01061505 GBYX01135134 JAO94787.1 UFQT01000109 SSX20149.1 APCN01005397 APCN01005398 APCN01005399 CH940650 EDW67478.2 CH902617 EDV42438.2 HAEB01019773 SBQ66300.1 CVRI01000006 CRK88153.1 CH964272 EDW84134.2 HADZ01016734 SBP80675.1 HAEE01001817 SBR21837.1 GFDL01011531 JAV23514.1 GFDL01011516 JAV23529.1 CH933806 EDW14468.2 JXUM01090538 KQ563846 KXJ73176.1 HAEC01013444 SBQ81661.1 HAEC01008141 SBQ76279.1 GAMC01010417 JAB96138.1 HAEJ01007273 SBS47730.1 JXUM01133534 KQ567992 KXJ69205.1 CM000070 EAL27887.4 AERX01048904 AERX01048905 AERX01048906 AERX01048907 AERX01048908 AERX01048909 AERX01048910 AERX01048911 AERX01048912 AERX01048913 JXJN01016991 CH477745 EAT36683.1 AE014297 ACL83574.2 AGB96429.1 AGB96427.1 AGB96428.1 AAF56821.5 GDHF01008033 JAI44281.1
Proteomes
UP000053268
UP000218220
UP000283053
UP000053240
UP000007266
UP000007755
+ More
UP000078540 UP000005203 UP000075809 UP000005205 UP000078541 UP000279307 UP000078492 UP000192223 UP000053105 UP000215335 UP000027135 UP000242457 UP000053097 UP000053825 UP000002358 UP000015103 UP000079169 UP000009046 UP000037069 UP000092553 UP000078542 UP000092462 UP000095301 UP000075840 UP000075920 UP000008792 UP000007801 UP000183832 UP000007798 UP000085678 UP000009192 UP000069940 UP000249989 UP000192220 UP000091820 UP000192221 UP000075885 UP000078200 UP000092445 UP000076408 UP000001819 UP000005207 UP000092460 UP000008820 UP000000803 UP000069272 UP000264800 UP000075881 UP000002852
UP000078540 UP000005203 UP000075809 UP000005205 UP000078541 UP000279307 UP000078492 UP000192223 UP000053105 UP000215335 UP000027135 UP000242457 UP000053097 UP000053825 UP000002358 UP000015103 UP000079169 UP000009046 UP000037069 UP000092553 UP000078542 UP000092462 UP000095301 UP000075840 UP000075920 UP000008792 UP000007801 UP000183832 UP000007798 UP000085678 UP000009192 UP000069940 UP000249989 UP000192220 UP000091820 UP000192221 UP000075885 UP000078200 UP000092445 UP000076408 UP000001819 UP000005207 UP000092460 UP000008820 UP000000803 UP000069272 UP000264800 UP000075881 UP000002852
Interpro
Gene 3D
ProteinModelPortal
A0A194Q830
A0A2A4JZT5
A0A3S2PIN2
A0A194QWX7
A0A139WBU9
F4W7D7
+ More
A0A195ATG9 A0A0C9RFH9 A0A0C9QTT3 V5H0L9 A0A088AIJ0 A0A151X8N0 A0A158NPV7 A0A151JT19 A0A1Y1KUL2 A0A1Y1KUK7 A0A3L8D806 A0A195DSL5 A0A1Y1KX53 A0A1B6BYQ6 A0A1B6CEC0 A0A1Y1KUM5 A0A1W4XHM2 A0A1B6J4N1 A0A1B6ILA6 A0A310SNE4 A0A1B6LK86 A0A2H8TWY4 A0A2S2PUB0 A0A0N0BGP5 A0A232FBC8 A0A2H8TGS0 A0A2S2R2L6 A0A067R3F9 A0A2A3EJQ7 A0A026WU37 A0A0L7RDF3 K7J6U9 A0A023F3N0 A0A0A9Y114 T1I6Z7 A0A146L604 A0A3Q0INC7 A0A0T6AWV2 E0VNL7 A0A1B6GXX3 A0A0L0BYF7 A0A0M4ENT6 A0A151IHE9 A0A1Q3G2A9 A0A224XV80 A0A1B0DHS1 A0A0S7LZL0 A0A1I8MCC3 A0A1I8MCB9 A0A336LU26 A0A182HYQ6 A0A182W3H5 A0A1S4G791 B4M167 B3M338 A0A1A8G3P1 A0A1J1HPL1 B4NKS3 A0A1A8CPG8 A0A1A8JPJ1 A0A1Q3F7G8 A0A1Q3F7M9 A0A1S3K6I7 B4K955 A0A182GV94 A0A2I4BUX1 A0A1A9WEG1 A0A1W4VU94 A0A1W4VTJ0 A0A182PP63 A0A1A9VAI0 A0A1A8HBV3 A0A1A8GYZ2 W8BGP2 A0A1A8UI27 A0A182HF49 A0A1B0A0G7 A0A182Y8Q9 Q298R8 I3J9T5 A0A1B0BML3 Q16QL0 B7Z0R2 A0A0B4KHD2 Q9VAS8 A0A0K8VZL8 A0A182FAM6 A0A3Q3GU39 A0A3Q3BAZ6 A0A182JRU7 A0A3B5RCJ7 A0A3B5Q0N2
A0A195ATG9 A0A0C9RFH9 A0A0C9QTT3 V5H0L9 A0A088AIJ0 A0A151X8N0 A0A158NPV7 A0A151JT19 A0A1Y1KUL2 A0A1Y1KUK7 A0A3L8D806 A0A195DSL5 A0A1Y1KX53 A0A1B6BYQ6 A0A1B6CEC0 A0A1Y1KUM5 A0A1W4XHM2 A0A1B6J4N1 A0A1B6ILA6 A0A310SNE4 A0A1B6LK86 A0A2H8TWY4 A0A2S2PUB0 A0A0N0BGP5 A0A232FBC8 A0A2H8TGS0 A0A2S2R2L6 A0A067R3F9 A0A2A3EJQ7 A0A026WU37 A0A0L7RDF3 K7J6U9 A0A023F3N0 A0A0A9Y114 T1I6Z7 A0A146L604 A0A3Q0INC7 A0A0T6AWV2 E0VNL7 A0A1B6GXX3 A0A0L0BYF7 A0A0M4ENT6 A0A151IHE9 A0A1Q3G2A9 A0A224XV80 A0A1B0DHS1 A0A0S7LZL0 A0A1I8MCC3 A0A1I8MCB9 A0A336LU26 A0A182HYQ6 A0A182W3H5 A0A1S4G791 B4M167 B3M338 A0A1A8G3P1 A0A1J1HPL1 B4NKS3 A0A1A8CPG8 A0A1A8JPJ1 A0A1Q3F7G8 A0A1Q3F7M9 A0A1S3K6I7 B4K955 A0A182GV94 A0A2I4BUX1 A0A1A9WEG1 A0A1W4VU94 A0A1W4VTJ0 A0A182PP63 A0A1A9VAI0 A0A1A8HBV3 A0A1A8GYZ2 W8BGP2 A0A1A8UI27 A0A182HF49 A0A1B0A0G7 A0A182Y8Q9 Q298R8 I3J9T5 A0A1B0BML3 Q16QL0 B7Z0R2 A0A0B4KHD2 Q9VAS8 A0A0K8VZL8 A0A182FAM6 A0A3Q3GU39 A0A3Q3BAZ6 A0A182JRU7 A0A3B5RCJ7 A0A3B5Q0N2
PDB
3A98
E-value=5.10395e-24,
Score=269
Ontologies
KEGG
GO
PANTHER
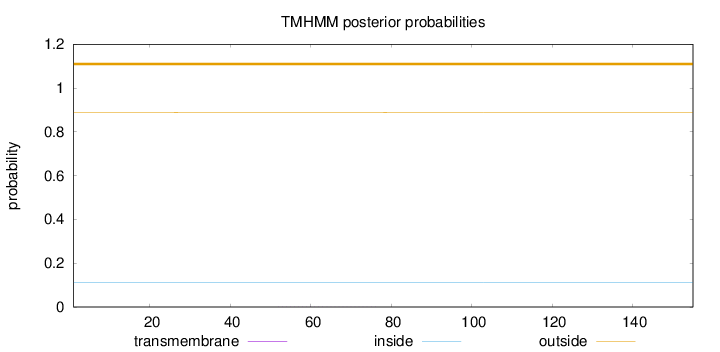
Topology
Length:
155
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00965
Exp number, first 60 AAs:
0.00384
Total prob of N-in:
0.11204
outside
1 - 155
Population Genetic Test Statistics
Pi
258.330815
Theta
182.992167
Tajima's D
1.34828
CLR
0.022904
CSRT
0.744112794360282
Interpretation
Uncertain
