Gene
KWMTBOMO12955
Pre Gene Modal
BGIBMGA000456
Annotation
PREDICTED:_uncharacterized_protein_LOC101744721_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.344
Sequence
CDS
ATGAGCTCTCCGGAGCCTCTGGTGCTCCAAGCGCTGCAGGGAAACACGCAGCTCAGTGACAAGTGGCCTGGACCTCGTGCCGTCAAGATATTCGTCGCTTCCGTATACAATGATTTTCGAGAAGAAAGACGGCAGATCTTAGAAATAGTCGGACCTGAACTCCAGTCAACTTACGATGATCGATACATCGAGATTGATTTTGTGGATATGCATTACGGAACAGACGGTGGAGATGAAACGAATCCGGCGTTATTACGGTATTATCTGGACGAAATAAGGTCTTGTAATCATTCCTCCAAAGCTGGATATTTTGTTTGTCTCTTTGGTGGTGATGCCTCATTGTACCACCCAGTTTTGCCTTATAAGATACCGTCAGATATCTTCGAACAGATGGTCATATCAGAAACAGCACAGGCTGACCTTGTTCGCGCCTGTTACCGTCTTAGTAGTGACGGGATTTACCACCTGGAAGACGATGACAAATGGTTCAGCGATCTTTTGGAACGAGACGAGCAACGTAGCCGCCTAGCCGAGGTACAGAAGGCATTCAATGCTGTGATTCTTGAGGCAATTTCCAGGGGAGTCGACGTTTCAAATCTGCTGCGAAGCCCAGTCGAAGCACAATGCGAATTAGCTCTGGAACTTTTATCAAACGGGAATCATCCCAAAGGACTTATAGCTGTGTTTCGTGACTTGGCGGAAGTTGAAGAGGACGACTCCAAAATTACTGTCCTCGCACATACAAGGCTGTTAGAACTGAAGAAAAAAATGGAAGAGACATTACCTGATACTCACTTTATAAGACTAGAGCCATCGTCTGCTGATTCTTCGAGGACGGATGGGGGATCTGAGAATGAAGAAAAATTAGCACCATTCAGGGAAAAAATTCTAGGCTTAGTGAGTTCACTAGTCGATGATAGTCTCAGTACTGAACCTGATCAAGGAAAGGGACGGAAGAAGACTGTACAGGAAGTTTTTCTCGAGCACATCACTCATTTAAGAATATGTATAGAACACGATCGAGCTTACAAAGTCAACGCTAAACAAATAGAAGAAGCATCTAAAAATATTTTGAGCACTGTGAAGGAAAATTACGAGAACCGAACTCGACATCCTCCGGTGTTAATATACGGGCCCGACGCTTCCGGAAAGACCACTTTGTTGACGCACTTGTACACAAAATGCGAGGAAATTTTCTCGAAGCCAGTTCTAAGGATAATAAGGTTTTCCGCTTCGACGCCGCGATCGGCCTACAACTTGGAACTTTTAAGAGTCATGTGCCAACAAATATCAATAATCTTGAACATTCCCGAAGGCTATTTACCTAAAGACGCCAGCTTTGATCCTCTGTACATTAATAACTGGTTTCAAAGTCTATTGCGGAGATGCGAAGAGATGGAAAATGAAATATTGTTTATATTTATTGATAACGTTCACAGAGTCAATCCTTTAGAGTGCGATATAGTAACAGGGCTGTCGTGGCTACCGATGTCTCTTCCAAGGAATGTCTTCCTAGTATGCACCAGTGCGATGTCTTTAGAGCAATTGCAATTGACCCCGGCGCAAAAGGAAAAGTTCAAAGTTCAGAACTGCTATTACTTTTTGGATGCTATCGAAGAAAGCCCGGAAAACAAATGCTACGGAGACTACATTGACGGTGCCTTCGATAATTTAGAAGTGGTCTTCGGTTCAAAGGCTTTTAGTAAATTAGCAAGTTATATAACTTGTTCGGAGTTTGGCCTAACGGAATTGGAACTTTTAGAACTTTTGATGCCGACGAGCAATAGCGAAGCCGTTATAACATTAAAAGATGCAAATTTTAACTTTTCAACGTTATGTGTCGCCAAGTACATGATGAAGAATCTCATAACAGAGACAATAGTGTCGGGACGTAGTACCTGGCGTTGGGCGGCGGCGGCGGCCGGTGCCCGGGCCCGACGCCGCTATGTTCGCGTGCAGAGCGCTCTTCGGGACGCCCATTCAGACCTTGCTGCACTACACTTCTCCTCCTTCCTCAACGACGCCGACGACACGGACACTTCAGAAGCACACGAGCTCGGTTGTGTAGACGATGACGACGCTCTTCTAGATTCGACGCCATTCCATTCGGCAAGTAGGACGGCGGCCGCTTTCACACAACGTCACGTAGAGGAGTCTTGGCTCCATTTACTTTTGGCTGGGGACTTCTCTAAGCTGAAGGACCTAACCGTTTGCAACTTCGACTTTTTACTAGCGGCCGTCCAAACGGTGACAATATCGTATTTACGCTGCATACTCGAGCACGTGCGTTGTTACATACTGGACAGAGACGTGGAACTCGTCTACGGAGCAGTTAAGAAGTCAAGTGACATATTAACACGGGATCCTATGCAGTTGGGAGCGCAGATAATAGCTTGGTTGCGGCCGGCAGTGGCTCGACGCGGTGTGCTCGCCACACTCGTGACAGCTGCGATGGCTTGGTGCGATGGATACGATAAGCCTCTACTCGTCCCTCTAAACGGCTGGCTTCACCCTCCTATAGCATCGACAGTCCGTGTCGTATCAGTGGGGGGCTCAAGTCCCGGTGCTGGTACAAGACTCCTCCAGTTGGCTCCCTCAGGACAGCATCTGGTACTCGCACCGTCGGCTGGAGACCCTCAACTTTGGCACATCATGTCGAATTCAAAGGTGCACACTTTCAAAGGACATTCGGGCAGCATTCTCTGTATGTGTGTGACCAGAGAATCACAATATCTACTCACAGGCTCTGAAGATACAAGCGTCATCGTCTGGGACTTACACACTTTAGCAGTTAAAACCAAAATATTAGAGCACATCGCACCTGTACTGTGCGTCGCAGCGATTGTGAATCGTTCGCTGGTGATTAGCGGTGGAGATGATTCCGCTGTTATAATCACATCTTTAGTGGACGGTGCCTTGGTAACAAAACTAGATCACCACAGAGGATCAGTGACATCTGTGAAAGTTATTCAAGATGGTAATATATTTATAACCGGATCCCAAGACGGCACTGTCTGCACTTGGAATGTAGAGAATTTTACCTTGCTATCAACTGTCACTGCTGGAGTGCCTATTCATATCTTCGATGTCACCGACGACAATGTCTTCCTACTCACACTGCAAGGGGACAACGAACTGCATCTTAGGACGTTCATAACGGGGACTTATTTACATCCCTTGAAAAGGCACAAAGCCAAGGTAAAGTGCTTCTGTGTTGCTCATGACTCGTCCCGGGTAGCGGTCGGTTGCGCCGACCAACGCATATATTTATATAGCTTACACAGCGCCCAACTGCTCCGGACATTGGCTGTGGCACACGACTTAGCAGCATTAGCTGTTGCTGATCGAGACCATTTCTTATTGGCTGCTGGTGGAAACAGAGTTACGATATACTCATTTCATACCGAAGACAACCTGACTAATTTCAGACCAACAAAACAAATGAAGAGACGTCAAACTAAATCCACCACAAATATTACTTTGCTTCAGGCTGAGCAAAGCGACTTGATACCAATAAGCTGCCTTGAAGTATCCCGAGACGGACAGCTTGGCGCCAGTGGATGCGCTCGTGGCCTTGTCAGAGTCTGGCAACTCTCAACACACAGATTACAAGCAACACTGAACGGCCATATAGGTCATATAACTTGTGTCACCTTTTCACCAAACAATCTATTGGTACTGAGTGGATCTGAAGACCGAACTGTGGTCGTTTGGCAGCTTGCGGATAATTCTCCTTCACTGACATATAAGGGTCATAGCTCAGCAATTCAAACCCTGCTGATGATGTCGGACGGGCGCCGTGCAATGTCAAGTGACAGGGCTCGTAATGTCCACGTCTGGCTCGTCGACTCTGGTATAGTTCTACATTCTGCAACCGGACCGTCAGCTAGTATTGACGTCACTTTAAATATGAAGTATGCTGTTCTATCGGACGGTGATAACTCAGTGCGGATCTGGTCTCTGGCTGGAGGTGATAATTCAGAAGACAACAGACGCTCAGTATCGCACGCCGAAAGAATCACCTGTTTCGCTTTGACCGCGGACTCACAGCATGTCGTCACTGGATCGATGGATATGTCACTGAAAGTTTGGAAATTGGATGGAGGGAAATTGTCACAGGTATTAGTCGGCCACTCAGACATAGTTACGTGCGTAGCAGTATCAATAACGAACAAAACTCAAGTTGTATCCGGCTCCTGGGACTGCAACCTTATTGTGTGGGACATAAATACTGGTTCTGATCTGCATTTGTTATCTGGACATCTTGGGAAAGTGACTTGCGTCAAAGTAACCGGGGACGGTACGATTGCGGTATCCTGCGCTGAAGACAAGACTCTTATAATATGGGAAACTAAAAGAGGTCTTGCTTTGACATCATTTTCATTACATGTCCCTTTACTCGGATTCCAAATAACAAGCGACTGTGCGAGAGTTGTAGTTCATCTTTTGGATAGAGGTTGTCTACCAATTATATGTTTGCACAATACACCGGCAACTTACGTTAAAATACCAACATATACAGCGCCAACCAAAGACGTGGATGAATTGCGTCCGTTGGCACCAAAAAGACCTATGAGAAGGCTACTGAAAAAGGAGGTATCACTTGATACGTACACGTGGCAGAAGAAATACGGACATCTCACTTCTGCGGCTATGATGGCTCAAGTAGATGAACGATTAAAAAGAAGATTCTCAGTATCTGCCTCTATGGAAGAAATATCCAAAATACAGGAGGCCAAGAATAAGGATTTGGGTTCACAGGTGAGTCTTGGTCCTGAGGAAGCAGCAATTGCTCAATCGCAACATTTCGATCAATTAGAAGCTTTGTGGAACAAGATATCACCTCCAAGACGTCGCTCAAATAAGTCCTTATCAAAGCAAAGTTCATTGATCGAAAGATTTGACTCTTCAGATGAAGAACATACCCCTGTCGAAGAACAAGAACATATGGTTGAGTAA
Protein
MSSPEPLVLQALQGNTQLSDKWPGPRAVKIFVASVYNDFREERRQILEIVGPELQSTYDDRYIEIDFVDMHYGTDGGDETNPALLRYYLDEIRSCNHSSKAGYFVCLFGGDASLYHPVLPYKIPSDIFEQMVISETAQADLVRACYRLSSDGIYHLEDDDKWFSDLLERDEQRSRLAEVQKAFNAVILEAISRGVDVSNLLRSPVEAQCELALELLSNGNHPKGLIAVFRDLAEVEEDDSKITVLAHTRLLELKKKMEETLPDTHFIRLEPSSADSSRTDGGSENEEKLAPFREKILGLVSSLVDDSLSTEPDQGKGRKKTVQEVFLEHITHLRICIEHDRAYKVNAKQIEEASKNILSTVKENYENRTRHPPVLIYGPDASGKTTLLTHLYTKCEEIFSKPVLRIIRFSASTPRSAYNLELLRVMCQQISIILNIPEGYLPKDASFDPLYINNWFQSLLRRCEEMENEILFIFIDNVHRVNPLECDIVTGLSWLPMSLPRNVFLVCTSAMSLEQLQLTPAQKEKFKVQNCYYFLDAIEESPENKCYGDYIDGAFDNLEVVFGSKAFSKLASYITCSEFGLTELELLELLMPTSNSEAVITLKDANFNFSTLCVAKYMMKNLITETIVSGRSTWRWAAAAAGARARRRYVRVQSALRDAHSDLAALHFSSFLNDADDTDTSEAHELGCVDDDDALLDSTPFHSASRTAAAFTQRHVEESWLHLLLAGDFSKLKDLTVCNFDFLLAAVQTVTISYLRCILEHVRCYILDRDVELVYGAVKKSSDILTRDPMQLGAQIIAWLRPAVARRGVLATLVTAAMAWCDGYDKPLLVPLNGWLHPPIASTVRVVSVGGSSPGAGTRLLQLAPSGQHLVLAPSAGDPQLWHIMSNSKVHTFKGHSGSILCMCVTRESQYLLTGSEDTSVIVWDLHTLAVKTKILEHIAPVLCVAAIVNRSLVISGGDDSAVIITSLVDGALVTKLDHHRGSVTSVKVIQDGNIFITGSQDGTVCTWNVENFTLLSTVTAGVPIHIFDVTDDNVFLLTLQGDNELHLRTFITGTYLHPLKRHKAKVKCFCVAHDSSRVAVGCADQRIYLYSLHSAQLLRTLAVAHDLAALAVADRDHFLLAAGGNRVTIYSFHTEDNLTNFRPTKQMKRRQTKSTTNITLLQAEQSDLIPISCLEVSRDGQLGASGCARGLVRVWQLSTHRLQATLNGHIGHITCVTFSPNNLLVLSGSEDRTVVVWQLADNSPSLTYKGHSSAIQTLLMMSDGRRAMSSDRARNVHVWLVDSGIVLHSATGPSASIDVTLNMKYAVLSDGDNSVRIWSLAGGDNSEDNRRSVSHAERITCFALTADSQHVVTGSMDMSLKVWKLDGGKLSQVLVGHSDIVTCVAVSITNKTQVVSGSWDCNLIVWDINTGSDLHLLSGHLGKVTCVKVTGDGTIAVSCAEDKTLIIWETKRGLALTSFSLHVPLLGFQITSDCARVVVHLLDRGCLPIICLHNTPATYVKIPTYTAPTKDVDELRPLAPKRPMRRLLKKEVSLDTYTWQKKYGHLTSAAMMAQVDERLKRRFSVSASMEEISKIQEAKNKDLGSQVSLGPEEAAIAQSQHFDQLEALWNKISPPRRRSNKSLSKQSSLIERFDSSDEEHTPVEEQEHMVE
Summary
Uniprot
A0A2A4JTT4
A0A3S2NJD8
A0A194Q7Q4
A0A212EYG3
A0A194QWF7
H9IT77
+ More
A0A2J7Q7K5 A0A182PUM3 Q7Q938 A0A182QE02 A0A084W9M3 Q17LP9 A0A182HXV6 A0A1L8DGI8 A0A182YBY0 A0A182WMH9 D6W857 A0A1J1J816 N6W4G1 Q28YR3 A0A0R1DSR6 A0A1W4VQT5 A0A1I8QF54 N6V8B0 A0A0R1DU17 A0A182NV14 A0A1I8QF96 A0A0R1DS00 B4P4D5 A0A1I8QF85 A0A0Q5VYD0 A0A1W4VQ32 A0A1I8QFD4 A0A0R1DS75 A0A0A1XAA7 A0A3B0JXN7 Q0E930 B3NMT8 A0A182JV88 A0A067R0Q3 A1ZB92 A0A1W4VD28 A0A0Q5VV14 A1ZB93 A0A2J7Q7K3 A0A0J9RFP9 A0A0P8XNK7 B4NMX8 A0A0J9RFQ4 Q6AWF2 B3MDG2 A0A0P9ADV4 A0A0J9U5D4 A0A0J9RG08 A0A0P8XNI6 B4J4N8 A0A0J9U5C8 A0A0J9RFX6 A0A0J9RG12 A0A3B0J3D9 B4KMA0 A0A0R3NNU6 B4HNQ1 A0A1I8QF80 A0A0Q9XK51 B4LNI7 A0A1W4VDD0 A0A0Q9W615 A0A0Q9W6G6 A0A2M4CIE3 A0A2M4CHJ1 W5JLH4 A0A2M4CJE3 A0A0M4EBH4 A0A0J9RFP3 A0A0Q9XIY4 A0A182J649 B4QCG3 A0A195BX57 A0A158NEV6 A0A195CAJ0 A0A151J6I6 A0A195FLB9 A0A0J7L6N7 A0A151WSM6 E2A0D1 E2C9A6 A0A026W2W0 W8AJN1 W8AVX2 F4WJA1 T1HSR0 A0A1S4N2W1 W8AAP5 E0W3M6 A0A0C9RZV5 A0A310SGI1 A0A1A9YTC7
A0A2J7Q7K5 A0A182PUM3 Q7Q938 A0A182QE02 A0A084W9M3 Q17LP9 A0A182HXV6 A0A1L8DGI8 A0A182YBY0 A0A182WMH9 D6W857 A0A1J1J816 N6W4G1 Q28YR3 A0A0R1DSR6 A0A1W4VQT5 A0A1I8QF54 N6V8B0 A0A0R1DU17 A0A182NV14 A0A1I8QF96 A0A0R1DS00 B4P4D5 A0A1I8QF85 A0A0Q5VYD0 A0A1W4VQ32 A0A1I8QFD4 A0A0R1DS75 A0A0A1XAA7 A0A3B0JXN7 Q0E930 B3NMT8 A0A182JV88 A0A067R0Q3 A1ZB92 A0A1W4VD28 A0A0Q5VV14 A1ZB93 A0A2J7Q7K3 A0A0J9RFP9 A0A0P8XNK7 B4NMX8 A0A0J9RFQ4 Q6AWF2 B3MDG2 A0A0P9ADV4 A0A0J9U5D4 A0A0J9RG08 A0A0P8XNI6 B4J4N8 A0A0J9U5C8 A0A0J9RFX6 A0A0J9RG12 A0A3B0J3D9 B4KMA0 A0A0R3NNU6 B4HNQ1 A0A1I8QF80 A0A0Q9XK51 B4LNI7 A0A1W4VDD0 A0A0Q9W615 A0A0Q9W6G6 A0A2M4CIE3 A0A2M4CHJ1 W5JLH4 A0A2M4CJE3 A0A0M4EBH4 A0A0J9RFP3 A0A0Q9XIY4 A0A182J649 B4QCG3 A0A195BX57 A0A158NEV6 A0A195CAJ0 A0A151J6I6 A0A195FLB9 A0A0J7L6N7 A0A151WSM6 E2A0D1 E2C9A6 A0A026W2W0 W8AJN1 W8AVX2 F4WJA1 T1HSR0 A0A1S4N2W1 W8AAP5 E0W3M6 A0A0C9RZV5 A0A310SGI1 A0A1A9YTC7
Pubmed
26354079
22118469
19121390
12364791
24438588
17510324
+ More
25244985 18362917 19820115 15632085 17994087 17550304 23185243 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 22936249 18057021 20920257 23761445 21347285 20798317 24508170 24495485 21719571 20566863
25244985 18362917 19820115 15632085 17994087 17550304 23185243 25830018 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24845553 22936249 18057021 20920257 23761445 21347285 20798317 24508170 24495485 21719571 20566863
EMBL
NWSH01000624
PCG75186.1
RSAL01000021
RVE52486.1
KQ459324
KPJ01553.1
+ More
AGBW02011563 OWR46484.1 KQ461073 KPJ09812.1 BABH01013442 BABH01013443 BABH01013444 NEVH01017442 PNF24563.1 AAAB01008905 EAA09696.4 AXCN02001008 ATLV01021838 KE525324 KFB46917.1 CH477213 EAT47653.1 APCN01005356 GFDF01008518 JAV05566.1 KQ971307 EFA11188.2 CVRI01000075 CRL08528.1 CM000071 ENO01722.1 EAL25901.1 CM000158 KRJ99998.1 ENO01721.1 KRT02662.1 KRJ99997.1 KRJ99995.1 KRJ99999.1 EDW91621.1 CH954179 KQS62090.1 KRJ99996.1 GBXI01006461 JAD07831.1 OUUW01000001 SPP75848.1 AE013599 AAF57681.2 AAM68460.1 EDV55092.1 KK853024 KDR12325.1 AAF57680.2 KQS62091.1 AAM68461.1 PNF24562.1 CM002911 KMY94863.1 CH902619 KPU76200.1 CH964282 EDW85717.1 KMY94856.1 KMY94868.1 BT015296 AAT94525.1 EDV36410.1 KPU76199.1 KMY94860.1 KMY94861.1 KPU76198.1 CH916367 EDW02743.1 KMY94855.1 KMY94857.1 KMY94867.1 KMY94859.1 KMY94862.1 KMY94864.1 KMY94865.1 KMY94866.1 SPP75847.1 CH933808 EDW08764.2 KRT02663.1 CH480816 EDW48470.1 KRG04264.1 CH940648 EDW62167.1 KRF80476.1 KRF80475.1 KRF80477.1 KRF80478.1 GGFL01000827 MBW65005.1 GGFL01000616 MBW64794.1 ADMH02001080 ETN64158.1 GGFL01000830 MBW65008.1 CP012524 ALC40877.1 KMY94858.1 KRG04263.1 CM000362 EDX07687.1 KQ976401 KYM92541.1 ADTU01013240 ADTU01013241 KQ978023 KYM97889.1 KQ979851 KYN18829.1 KQ981490 KYN41057.1 LBMM01000514 KMQ98169.1 KQ982769 KYQ50823.1 GL435569 EFN73134.1 GL453806 EFN75451.1 KK107455 EZA50412.1 GAMC01021637 JAB84918.1 GAMC01021639 JAB84916.1 GL888182 EGI65679.1 ACPB03014480 AAZO01007425 GAMC01021635 JAB84920.1 DS235882 EEB20232.1 GBYB01013671 JAG83438.1 KQ760184 OAD61624.1
AGBW02011563 OWR46484.1 KQ461073 KPJ09812.1 BABH01013442 BABH01013443 BABH01013444 NEVH01017442 PNF24563.1 AAAB01008905 EAA09696.4 AXCN02001008 ATLV01021838 KE525324 KFB46917.1 CH477213 EAT47653.1 APCN01005356 GFDF01008518 JAV05566.1 KQ971307 EFA11188.2 CVRI01000075 CRL08528.1 CM000071 ENO01722.1 EAL25901.1 CM000158 KRJ99998.1 ENO01721.1 KRT02662.1 KRJ99997.1 KRJ99995.1 KRJ99999.1 EDW91621.1 CH954179 KQS62090.1 KRJ99996.1 GBXI01006461 JAD07831.1 OUUW01000001 SPP75848.1 AE013599 AAF57681.2 AAM68460.1 EDV55092.1 KK853024 KDR12325.1 AAF57680.2 KQS62091.1 AAM68461.1 PNF24562.1 CM002911 KMY94863.1 CH902619 KPU76200.1 CH964282 EDW85717.1 KMY94856.1 KMY94868.1 BT015296 AAT94525.1 EDV36410.1 KPU76199.1 KMY94860.1 KMY94861.1 KPU76198.1 CH916367 EDW02743.1 KMY94855.1 KMY94857.1 KMY94867.1 KMY94859.1 KMY94862.1 KMY94864.1 KMY94865.1 KMY94866.1 SPP75847.1 CH933808 EDW08764.2 KRT02663.1 CH480816 EDW48470.1 KRG04264.1 CH940648 EDW62167.1 KRF80476.1 KRF80475.1 KRF80477.1 KRF80478.1 GGFL01000827 MBW65005.1 GGFL01000616 MBW64794.1 ADMH02001080 ETN64158.1 GGFL01000830 MBW65008.1 CP012524 ALC40877.1 KMY94858.1 KRG04263.1 CM000362 EDX07687.1 KQ976401 KYM92541.1 ADTU01013240 ADTU01013241 KQ978023 KYM97889.1 KQ979851 KYN18829.1 KQ981490 KYN41057.1 LBMM01000514 KMQ98169.1 KQ982769 KYQ50823.1 GL435569 EFN73134.1 GL453806 EFN75451.1 KK107455 EZA50412.1 GAMC01021637 JAB84918.1 GAMC01021639 JAB84916.1 GL888182 EGI65679.1 ACPB03014480 AAZO01007425 GAMC01021635 JAB84920.1 DS235882 EEB20232.1 GBYB01013671 JAG83438.1 KQ760184 OAD61624.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000007151
UP000053240
UP000005204
+ More
UP000235965 UP000075885 UP000007062 UP000075886 UP000030765 UP000008820 UP000075840 UP000076408 UP000075920 UP000007266 UP000183832 UP000001819 UP000002282 UP000192221 UP000095300 UP000075884 UP000008711 UP000268350 UP000000803 UP000075881 UP000027135 UP000007801 UP000007798 UP000001070 UP000009192 UP000001292 UP000008792 UP000000673 UP000092553 UP000075880 UP000000304 UP000078540 UP000005205 UP000078542 UP000078492 UP000078541 UP000036403 UP000075809 UP000000311 UP000008237 UP000053097 UP000007755 UP000015103 UP000009046 UP000092443
UP000235965 UP000075885 UP000007062 UP000075886 UP000030765 UP000008820 UP000075840 UP000076408 UP000075920 UP000007266 UP000183832 UP000001819 UP000002282 UP000192221 UP000095300 UP000075884 UP000008711 UP000268350 UP000000803 UP000075881 UP000027135 UP000007801 UP000007798 UP000001070 UP000009192 UP000001292 UP000008792 UP000000673 UP000092553 UP000075880 UP000000304 UP000078540 UP000005205 UP000078542 UP000078492 UP000078541 UP000036403 UP000075809 UP000000311 UP000008237 UP000053097 UP000007755 UP000015103 UP000009046 UP000092443
Interpro
IPR001680
WD40_repeat
+ More
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR019775 WD40_repeat_CS
IPR020472 G-protein_beta_WD-40_rep
IPR017986 WD40_repeat_dom
IPR027417 P-loop_NTPase
IPR036322 WD40_repeat_dom_sf
IPR003593 AAA+_ATPase
IPR041664 AAA_16
IPR024977 Apc4_WD40_dom
IPR011044 Quino_amine_DH_bsu
IPR002372 PQQ_repeat
IPR025139 DUF4062
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR011047 Quinoprotein_ADH-like_supfam
IPR019775 WD40_repeat_CS
IPR020472 G-protein_beta_WD-40_rep
IPR017986 WD40_repeat_dom
IPR027417 P-loop_NTPase
IPR036322 WD40_repeat_dom_sf
IPR003593 AAA+_ATPase
IPR041664 AAA_16
IPR024977 Apc4_WD40_dom
IPR011044 Quino_amine_DH_bsu
IPR002372 PQQ_repeat
IPR025139 DUF4062
Gene 3D
ProteinModelPortal
A0A2A4JTT4
A0A3S2NJD8
A0A194Q7Q4
A0A212EYG3
A0A194QWF7
H9IT77
+ More
A0A2J7Q7K5 A0A182PUM3 Q7Q938 A0A182QE02 A0A084W9M3 Q17LP9 A0A182HXV6 A0A1L8DGI8 A0A182YBY0 A0A182WMH9 D6W857 A0A1J1J816 N6W4G1 Q28YR3 A0A0R1DSR6 A0A1W4VQT5 A0A1I8QF54 N6V8B0 A0A0R1DU17 A0A182NV14 A0A1I8QF96 A0A0R1DS00 B4P4D5 A0A1I8QF85 A0A0Q5VYD0 A0A1W4VQ32 A0A1I8QFD4 A0A0R1DS75 A0A0A1XAA7 A0A3B0JXN7 Q0E930 B3NMT8 A0A182JV88 A0A067R0Q3 A1ZB92 A0A1W4VD28 A0A0Q5VV14 A1ZB93 A0A2J7Q7K3 A0A0J9RFP9 A0A0P8XNK7 B4NMX8 A0A0J9RFQ4 Q6AWF2 B3MDG2 A0A0P9ADV4 A0A0J9U5D4 A0A0J9RG08 A0A0P8XNI6 B4J4N8 A0A0J9U5C8 A0A0J9RFX6 A0A0J9RG12 A0A3B0J3D9 B4KMA0 A0A0R3NNU6 B4HNQ1 A0A1I8QF80 A0A0Q9XK51 B4LNI7 A0A1W4VDD0 A0A0Q9W615 A0A0Q9W6G6 A0A2M4CIE3 A0A2M4CHJ1 W5JLH4 A0A2M4CJE3 A0A0M4EBH4 A0A0J9RFP3 A0A0Q9XIY4 A0A182J649 B4QCG3 A0A195BX57 A0A158NEV6 A0A195CAJ0 A0A151J6I6 A0A195FLB9 A0A0J7L6N7 A0A151WSM6 E2A0D1 E2C9A6 A0A026W2W0 W8AJN1 W8AVX2 F4WJA1 T1HSR0 A0A1S4N2W1 W8AAP5 E0W3M6 A0A0C9RZV5 A0A310SGI1 A0A1A9YTC7
A0A2J7Q7K5 A0A182PUM3 Q7Q938 A0A182QE02 A0A084W9M3 Q17LP9 A0A182HXV6 A0A1L8DGI8 A0A182YBY0 A0A182WMH9 D6W857 A0A1J1J816 N6W4G1 Q28YR3 A0A0R1DSR6 A0A1W4VQT5 A0A1I8QF54 N6V8B0 A0A0R1DU17 A0A182NV14 A0A1I8QF96 A0A0R1DS00 B4P4D5 A0A1I8QF85 A0A0Q5VYD0 A0A1W4VQ32 A0A1I8QFD4 A0A0R1DS75 A0A0A1XAA7 A0A3B0JXN7 Q0E930 B3NMT8 A0A182JV88 A0A067R0Q3 A1ZB92 A0A1W4VD28 A0A0Q5VV14 A1ZB93 A0A2J7Q7K3 A0A0J9RFP9 A0A0P8XNK7 B4NMX8 A0A0J9RFQ4 Q6AWF2 B3MDG2 A0A0P9ADV4 A0A0J9U5D4 A0A0J9RG08 A0A0P8XNI6 B4J4N8 A0A0J9U5C8 A0A0J9RFX6 A0A0J9RG12 A0A3B0J3D9 B4KMA0 A0A0R3NNU6 B4HNQ1 A0A1I8QF80 A0A0Q9XK51 B4LNI7 A0A1W4VDD0 A0A0Q9W615 A0A0Q9W6G6 A0A2M4CIE3 A0A2M4CHJ1 W5JLH4 A0A2M4CJE3 A0A0M4EBH4 A0A0J9RFP3 A0A0Q9XIY4 A0A182J649 B4QCG3 A0A195BX57 A0A158NEV6 A0A195CAJ0 A0A151J6I6 A0A195FLB9 A0A0J7L6N7 A0A151WSM6 E2A0D1 E2C9A6 A0A026W2W0 W8AJN1 W8AVX2 F4WJA1 T1HSR0 A0A1S4N2W1 W8AAP5 E0W3M6 A0A0C9RZV5 A0A310SGI1 A0A1A9YTC7
PDB
2YMU
E-value=4.755e-32,
Score=350
Ontologies
GO
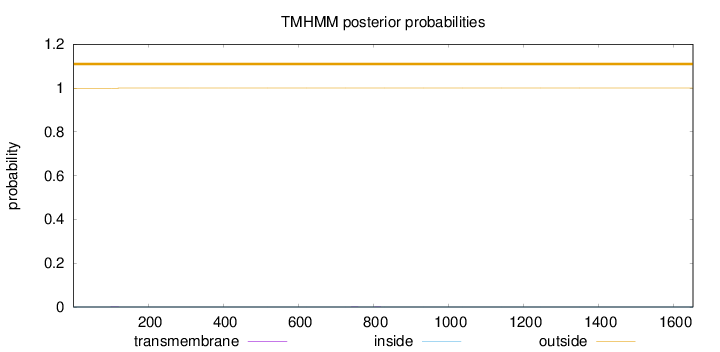
Topology
Length:
1652
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0187199999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00052
outside
1 - 1652
Population Genetic Test Statistics
Pi
29.381729
Theta
169.971069
Tajima's D
-1.288931
CLR
2252.316436
CSRT
0.088145592720364
Interpretation
Uncertain
