Gene
KWMTBOMO12950
Annotation
PREDICTED:_uncharacterized_protein_LOC105842415_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.183
Sequence
CDS
ATGAGTGAGAAAGAAGAAAAGGAGTTAGTTAAAAAACGAGCTAGTCTAAAAGGCCGATTGACTACATTCAGAAAGTACATCGAGGAATTGAGCGGTACGTTGAATCAGCAACAAGTAAATGAATTGCAATTAAGAATCGGTAAGATCGAATTATTATTTGAACAATATGACGTAGTACAAACTAAAATAGAATGCCTCGCTGACGATTTCAGCTCGCAAAGCACAGAGAGAGAAGATTTTGAATCAATTTATTACAAATCCCTTTCACAGGCTCAGGAGATTCTTTCTCGTAATTCTAAATTGTTACAAACCCAAGTGCATAGTTTTGAGGGTTGCTCTCATGTGAGTAATCATAGCCTGGTTAAATTGCCTACCATTAGTCTACCCAAATTTAATGGTTCATATAGCAACTGGCTGGAATTCCATGACACCTTCAGTAGTCTTATACATTCGAACGAGGCGATAGGAGAAATTAATAAATTCCACTACCTCAGGGCCTCGCTAGAAGGCTCTGCTGCTGTACTGCTTCAGGAGATTCAGTTTTCTGCCAGCAACTATTCTGTGGCGTGGAAGCTGTTATGTGAGAGGTACGATAATAAGCGCCTATTAATTCAAACACACGTTAATAATTTATTTAATGTAGACGCAATAAAGAGTGAATCTTCATTTAATTTAAGACGTTTCATAGATCAGCTGAATAAAAGCCTCAGAGCTTTAGAGTCGCTGGGTGAACCGACTGATCATTGGGACACTCTTCTCATCCACATGATGAGTCGTAAATTCGAACAGAAGACGCTGCGTGAATGGGAAGAGTGCAAAGGTCGTCTTGATAAAGAAAAGCCCATTTCTTTGTCCATCTTTATCGATTTCATTCGTAACCGGGCCGATCTATTAGAAACGATGGAATTTACTCGCCCTAACCTTCCTCACACACAAGTTCAGCCACCACAGTCGCCTAAAGGTCAGAAAATTAAAACATTAGTTTCTGTGTCCGGAACGCAGCCGTCGTCGAGTGGCGGCGCTAGTATTATTTCTGGTTCCGGTACAACATCGTGCCCACACTGTAAGGTTCAAGGTGTGTTACAACTGCTTTCGTAG
Protein
MSEKEEKELVKKRASLKGRLTTFRKYIEELSGTLNQQQVNELQLRIGKIELLFEQYDVVQTKIECLADDFSSQSTEREDFESIYYKSLSQAQEILSRNSKLLQTQVHSFEGCSHVSNHSLVKLPTISLPKFNGSYSNWLEFHDTFSSLIHSNEAIGEINKFHYLRASLEGSAAVLLQEIQFSASNYSVAWKLLCERYDNKRLLIQTHVNNLFNVDAIKSESSFNLRRFIDQLNKSLRALESLGEPTDHWDTLLIHMMSRKFEQKTLREWEECKGRLDKEKPISLSIFIDFIRNRADLLETMEFTRPNLPHTQVQPPQSPKGQKIKTLVSVSGTQPSSSGGASIISGSGTTSCPHCKVQGVLQLLS
Summary
Uniprot
A0A2A4JBY8
H9JIA8
H9J9C1
A0A0L7LB62
A0A0L7LCW4
A0A0L7L574
+ More
A0A0N1IBH6 A0A194RGQ8 A0A151IIA8 A0A0L7KRI4 A0A194RI04 A0A2A4JL65 V5GYU6 V5GPJ6 A0A2A4JLG7 A0A2H1WXG3 S4NIJ2 A0A0J7KD82 A0A2A4K5Q9 A0A194RAG7 V5I888 V5I8W0 A0A1Y1LJL9 A0A151IJK3 A0A151IDD4 A0A0A9Z2N1 A0A069DYA1 A0A3S2N9P3 A0A151X752 A0A1E1WLQ7 A0A151ICE7 A0A151IRH4 A0A2S2NIF1 A0A2S2PCB8 A0A151IMM1 A0A2S2P390 A0A195C7P8 A0A1Y1MWT5 A0A151ITI6 X1WJH1 A0A023EZB2 X1XU47 A0A151JAJ1 A0A151IIA5 A0A1Y1KHZ5 X1WVV7 A0A151XJI2 A0A182GTJ7 A0A2S2PTX9 A0A1Y1N4T1 A0A151IDJ5 A0A151XF58 A0A151IN79 A0A0L7QYE0 A0A194QJ48 A0A139W8E1 A0A0J7N4U0 A0A023EZC2 J9JT64 A0A1Y1KFT0 A0A2S2NTG1 A0A0N1IPF2 A0A2S2NH31 A0A2S2Q0A5 J9LHB7 X1X9I2 D7GYB7 A0A2S2NZD1 A0A151I8M6 J9LVB4 A0A151JNR8 X1WPD9 A0A151JQI5 A0A151JF33 A0A2S2PNZ4 J9LAN2 A0A154P493 A0A182RIS2 J9LYM7 A0A023EZK8 J9KCB6 X1X1J0 X1WL73 A0A2S2PLU2 A0A151IYW7 A0A151I689 A0A151IR94 A0A2S2PUJ2
A0A0N1IBH6 A0A194RGQ8 A0A151IIA8 A0A0L7KRI4 A0A194RI04 A0A2A4JL65 V5GYU6 V5GPJ6 A0A2A4JLG7 A0A2H1WXG3 S4NIJ2 A0A0J7KD82 A0A2A4K5Q9 A0A194RAG7 V5I888 V5I8W0 A0A1Y1LJL9 A0A151IJK3 A0A151IDD4 A0A0A9Z2N1 A0A069DYA1 A0A3S2N9P3 A0A151X752 A0A1E1WLQ7 A0A151ICE7 A0A151IRH4 A0A2S2NIF1 A0A2S2PCB8 A0A151IMM1 A0A2S2P390 A0A195C7P8 A0A1Y1MWT5 A0A151ITI6 X1WJH1 A0A023EZB2 X1XU47 A0A151JAJ1 A0A151IIA5 A0A1Y1KHZ5 X1WVV7 A0A151XJI2 A0A182GTJ7 A0A2S2PTX9 A0A1Y1N4T1 A0A151IDJ5 A0A151XF58 A0A151IN79 A0A0L7QYE0 A0A194QJ48 A0A139W8E1 A0A0J7N4U0 A0A023EZC2 J9JT64 A0A1Y1KFT0 A0A2S2NTG1 A0A0N1IPF2 A0A2S2NH31 A0A2S2Q0A5 J9LHB7 X1X9I2 D7GYB7 A0A2S2NZD1 A0A151I8M6 J9LVB4 A0A151JNR8 X1WPD9 A0A151JQI5 A0A151JF33 A0A2S2PNZ4 J9LAN2 A0A154P493 A0A182RIS2 J9LYM7 A0A023EZK8 J9KCB6 X1X1J0 X1WL73 A0A2S2PLU2 A0A151IYW7 A0A151I689 A0A151IR94 A0A2S2PUJ2
Pubmed
EMBL
NWSH01001945
PCG69607.1
BABH01022219
BABH01022220
BABH01004450
JTDY01001855
+ More
KOB72712.1 JTDY01001654 KOB73239.1 JTDY01006305 JTDY01002831 KOB66118.1 KOB70633.1 KQ459312 KPJ01687.1 KQ460211 KPJ16510.1 KQ977505 KYN02283.1 JTDY01006550 JTDY01002030 JTDY01001260 KOB65898.1 KOB72276.1 KOB74365.1 KQ460205 KPJ16960.1 NWSH01001201 PCG72150.1 GALX01002843 JAB65623.1 GALX01002427 JAB66039.1 NWSH01001093 PCG72659.1 ODYU01011804 SOQ57759.1 GAIX01014054 JAA78506.1 LBMM01009058 KMQ88428.1 NWSH01000142 PCG79113.1 KQ460500 KPJ14270.1 GALX01005110 JAB63356.1 GALX01004000 JAB64466.1 GEZM01055231 JAV73081.1 KQ977342 KYN03430.1 KQ977963 KYM98425.1 GBHO01005506 GBHO01005503 JAG38098.1 JAG38101.1 GBGD01000063 JAC88826.1 RSAL01000402 RVE41913.1 KQ982464 KYQ56128.1 GDQN01003166 JAT87888.1 KQ978056 KYM97755.1 KQ981127 KYN09299.1 GGMR01004354 MBY16973.1 GGMR01014413 MBY27032.1 KQ977032 KYN06203.1 GGMR01011310 MBY23929.1 KQ978164 KYM96665.1 GEZM01018702 GEZM01018700 GEZM01018697 JAV90049.1 KQ981002 KYN10407.1 ABLF02021176 ABLF02066403 GBBI01004140 JAC14572.1 ABLF02006001 ABLF02006002 KQ979276 KYN22031.1 KQ977557 KYN02007.1 GEZM01086714 JAV59026.1 ABLF02041505 KQ982064 KYQ60564.1 JXUM01252006 KQ640254 KXJ62375.1 GGMR01020322 MBY32941.1 GEZM01012999 JAV92824.1 KQ977959 KYM98432.1 KQ982204 KYQ59009.1 KQ976954 KYN06860.1 KQ414692 KOC63604.1 KQ458793 KPJ04955.1 KQ973442 KXZ75531.1 LBMM01010061 KMQ87695.1 GBBI01004139 JAC14573.1 ABLF02023842 ABLF02028949 ABLF02058391 GEZM01086715 JAV59020.1 GGMR01007417 MBY20036.1 KQ460545 KPJ14069.1 GGMR01003885 MBY16504.1 GGMS01001896 MBY71099.1 ABLF02022040 ABLF02056888 ABLF02042264 GG695382 EFA13346.1 GGMR01009911 MBY22530.1 KQ978343 KYM95005.1 ABLF02004142 KQ978855 KYN27942.1 ABLF02006985 KQ978679 KYN29321.1 KQ979016 KYN24380.1 GGMR01018506 MBY31125.1 ABLF02013385 ABLF02029502 ABLF02029511 KQ434810 KZC06736.1 ABLF02016109 ABLF02016113 GBBI01004032 JAC14680.1 ABLF02026305 GGMR01017803 MBY30422.1 KQ980737 KYN13736.1 KQ978502 KYM93420.1 KQ981142 KYN09177.1 GGMR01020500 MBY33119.1
KOB72712.1 JTDY01001654 KOB73239.1 JTDY01006305 JTDY01002831 KOB66118.1 KOB70633.1 KQ459312 KPJ01687.1 KQ460211 KPJ16510.1 KQ977505 KYN02283.1 JTDY01006550 JTDY01002030 JTDY01001260 KOB65898.1 KOB72276.1 KOB74365.1 KQ460205 KPJ16960.1 NWSH01001201 PCG72150.1 GALX01002843 JAB65623.1 GALX01002427 JAB66039.1 NWSH01001093 PCG72659.1 ODYU01011804 SOQ57759.1 GAIX01014054 JAA78506.1 LBMM01009058 KMQ88428.1 NWSH01000142 PCG79113.1 KQ460500 KPJ14270.1 GALX01005110 JAB63356.1 GALX01004000 JAB64466.1 GEZM01055231 JAV73081.1 KQ977342 KYN03430.1 KQ977963 KYM98425.1 GBHO01005506 GBHO01005503 JAG38098.1 JAG38101.1 GBGD01000063 JAC88826.1 RSAL01000402 RVE41913.1 KQ982464 KYQ56128.1 GDQN01003166 JAT87888.1 KQ978056 KYM97755.1 KQ981127 KYN09299.1 GGMR01004354 MBY16973.1 GGMR01014413 MBY27032.1 KQ977032 KYN06203.1 GGMR01011310 MBY23929.1 KQ978164 KYM96665.1 GEZM01018702 GEZM01018700 GEZM01018697 JAV90049.1 KQ981002 KYN10407.1 ABLF02021176 ABLF02066403 GBBI01004140 JAC14572.1 ABLF02006001 ABLF02006002 KQ979276 KYN22031.1 KQ977557 KYN02007.1 GEZM01086714 JAV59026.1 ABLF02041505 KQ982064 KYQ60564.1 JXUM01252006 KQ640254 KXJ62375.1 GGMR01020322 MBY32941.1 GEZM01012999 JAV92824.1 KQ977959 KYM98432.1 KQ982204 KYQ59009.1 KQ976954 KYN06860.1 KQ414692 KOC63604.1 KQ458793 KPJ04955.1 KQ973442 KXZ75531.1 LBMM01010061 KMQ87695.1 GBBI01004139 JAC14573.1 ABLF02023842 ABLF02028949 ABLF02058391 GEZM01086715 JAV59020.1 GGMR01007417 MBY20036.1 KQ460545 KPJ14069.1 GGMR01003885 MBY16504.1 GGMS01001896 MBY71099.1 ABLF02022040 ABLF02056888 ABLF02042264 GG695382 EFA13346.1 GGMR01009911 MBY22530.1 KQ978343 KYM95005.1 ABLF02004142 KQ978855 KYN27942.1 ABLF02006985 KQ978679 KYN29321.1 KQ979016 KYN24380.1 GGMR01018506 MBY31125.1 ABLF02013385 ABLF02029502 ABLF02029511 KQ434810 KZC06736.1 ABLF02016109 ABLF02016113 GBBI01004032 JAC14680.1 ABLF02026305 GGMR01017803 MBY30422.1 KQ980737 KYN13736.1 KQ978502 KYM93420.1 KQ981142 KYN09177.1 GGMR01020500 MBY33119.1
Proteomes
Pfam
Interpro
IPR036397
RNaseH_sf
+ More
IPR005312 DUF1759
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR001878 Znf_CCHC
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041588 Integrase_H2C2
IPR021109 Peptidase_aspartic_dom_sf
IPR001995 Peptidase_A2_cat
IPR003121 SWIB_MDM2_domain
IPR036885 SWIB_MDM2_dom_sf
IPR019835 SWIB_domain
IPR001969 Aspartic_peptidase_AS
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR013098 Ig_I-set
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR038563 Endonuclease_7_sf
IPR004211 Endonuclease_7
IPR005312 DUF1759
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR001878 Znf_CCHC
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041588 Integrase_H2C2
IPR021109 Peptidase_aspartic_dom_sf
IPR001995 Peptidase_A2_cat
IPR003121 SWIB_MDM2_domain
IPR036885 SWIB_MDM2_dom_sf
IPR019835 SWIB_domain
IPR001969 Aspartic_peptidase_AS
IPR003598 Ig_sub2
IPR036179 Ig-like_dom_sf
IPR013098 Ig_I-set
IPR003599 Ig_sub
IPR007110 Ig-like_dom
IPR013783 Ig-like_fold
IPR038563 Endonuclease_7_sf
IPR004211 Endonuclease_7
ProteinModelPortal
A0A2A4JBY8
H9JIA8
H9J9C1
A0A0L7LB62
A0A0L7LCW4
A0A0L7L574
+ More
A0A0N1IBH6 A0A194RGQ8 A0A151IIA8 A0A0L7KRI4 A0A194RI04 A0A2A4JL65 V5GYU6 V5GPJ6 A0A2A4JLG7 A0A2H1WXG3 S4NIJ2 A0A0J7KD82 A0A2A4K5Q9 A0A194RAG7 V5I888 V5I8W0 A0A1Y1LJL9 A0A151IJK3 A0A151IDD4 A0A0A9Z2N1 A0A069DYA1 A0A3S2N9P3 A0A151X752 A0A1E1WLQ7 A0A151ICE7 A0A151IRH4 A0A2S2NIF1 A0A2S2PCB8 A0A151IMM1 A0A2S2P390 A0A195C7P8 A0A1Y1MWT5 A0A151ITI6 X1WJH1 A0A023EZB2 X1XU47 A0A151JAJ1 A0A151IIA5 A0A1Y1KHZ5 X1WVV7 A0A151XJI2 A0A182GTJ7 A0A2S2PTX9 A0A1Y1N4T1 A0A151IDJ5 A0A151XF58 A0A151IN79 A0A0L7QYE0 A0A194QJ48 A0A139W8E1 A0A0J7N4U0 A0A023EZC2 J9JT64 A0A1Y1KFT0 A0A2S2NTG1 A0A0N1IPF2 A0A2S2NH31 A0A2S2Q0A5 J9LHB7 X1X9I2 D7GYB7 A0A2S2NZD1 A0A151I8M6 J9LVB4 A0A151JNR8 X1WPD9 A0A151JQI5 A0A151JF33 A0A2S2PNZ4 J9LAN2 A0A154P493 A0A182RIS2 J9LYM7 A0A023EZK8 J9KCB6 X1X1J0 X1WL73 A0A2S2PLU2 A0A151IYW7 A0A151I689 A0A151IR94 A0A2S2PUJ2
A0A0N1IBH6 A0A194RGQ8 A0A151IIA8 A0A0L7KRI4 A0A194RI04 A0A2A4JL65 V5GYU6 V5GPJ6 A0A2A4JLG7 A0A2H1WXG3 S4NIJ2 A0A0J7KD82 A0A2A4K5Q9 A0A194RAG7 V5I888 V5I8W0 A0A1Y1LJL9 A0A151IJK3 A0A151IDD4 A0A0A9Z2N1 A0A069DYA1 A0A3S2N9P3 A0A151X752 A0A1E1WLQ7 A0A151ICE7 A0A151IRH4 A0A2S2NIF1 A0A2S2PCB8 A0A151IMM1 A0A2S2P390 A0A195C7P8 A0A1Y1MWT5 A0A151ITI6 X1WJH1 A0A023EZB2 X1XU47 A0A151JAJ1 A0A151IIA5 A0A1Y1KHZ5 X1WVV7 A0A151XJI2 A0A182GTJ7 A0A2S2PTX9 A0A1Y1N4T1 A0A151IDJ5 A0A151XF58 A0A151IN79 A0A0L7QYE0 A0A194QJ48 A0A139W8E1 A0A0J7N4U0 A0A023EZC2 J9JT64 A0A1Y1KFT0 A0A2S2NTG1 A0A0N1IPF2 A0A2S2NH31 A0A2S2Q0A5 J9LHB7 X1X9I2 D7GYB7 A0A2S2NZD1 A0A151I8M6 J9LVB4 A0A151JNR8 X1WPD9 A0A151JQI5 A0A151JF33 A0A2S2PNZ4 J9LAN2 A0A154P493 A0A182RIS2 J9LYM7 A0A023EZK8 J9KCB6 X1X1J0 X1WL73 A0A2S2PLU2 A0A151IYW7 A0A151I689 A0A151IR94 A0A2S2PUJ2
Ontologies
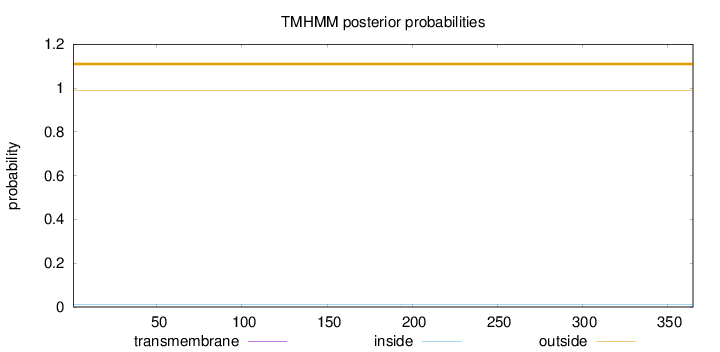
Topology
Length:
365
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00107
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00993
outside
1 - 365
Population Genetic Test Statistics
Pi
245.840863
Theta
215.471336
Tajima's D
0.43656
CLR
0
CSRT
0.493125343732813
Interpretation
Uncertain
