Gene
KWMTBOMO12947 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000236
Annotation
isovaleryl_coenzyme_A_dehydrogenase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.522 Mitochondrial Reliability : 2.024
Sequence
CDS
ATGGTTGTTAGGTTGTGTGTGAGACGCTTGCTAAGGAAAACCGCGAATAAAACCGGTCATCGATGCATCTCGCATTATCCCATTGACGATCATGTGTTTGGACTCTCCGAAGAACAACAACAGCTCAGAAAAATGGTATTCGACTTTGCTCAGAAGGAATTGGCGCCAAAAGCAGCTGAAATCGACAAAGAAAACAATTTTAAAGAACTAAGGCCATTTTGGAAGAAACTTGGAGATCTCGGATTGCTAGGTATTACCGCAAGTTCCGACTATGGAGGTACAGGGGGAAAATATTCGGACCATTGTGTCATTATGGAGGAATTATCAAGGGCCAGCGGTGGTATCGCCTTGTCATATGGAGCTCATTCGAATTTGTGCGTGAACCAGATCAACAGAAACGGTACAGAGGAGCAGAAAAGTAAATATTTACCAAAGTTATGTTCAGGAGAGCATATCGGAGCACTGGCAATGTCCGAACCAGGGTCAGGCAGTGACGTGGTGTCTATGAAGTTACGAGCAGAGAAGAAAGGCGATTATTATGTGTTGAACGGAAATAAGTTTTGGATTACTAATGGACCAGACGCAGATGTTTTAGTGGTATACGCCAAGACAAATTGGTCGACGAGTAAACAACAACATGGGATTTCAGCTTTTTTAATTGAAAAAGATTATCCTGGGTTTTCAACAGCTCAAAAATTAGATAAACTAGGTATGAGGGGTTCAAACACAGGGGAATTGGTGTTTGAGGATTGTAAGGTACCGGCAGCAAATTTATTAGGACAGGAAAACAAAGGGGTGTACGTACTAATGTCCGGCTTAGACCTGGAGAGACTGGTTCTTGCTGCAGGTCCCGTCGGATTAATGCAGGCCGCGATAGACACAGCCTTCTTGTACGCACACACCAGGAAACAATTTGGGAAGAACATTGGAGAGTTTCAATTGATACAGGGTAAAATGGCAGATATGTATACAACGTTAAGTGCATGTCGTAGCTACTTATACAATGTGGCTAAAGCGTGTGATAACGGCCATGTTAACAGTAAGGATTGCGCGGGCGTGATACTCTACTGTGCGGAAAAAGCAACACAGGTCGCTTTAGATGCAATCCAAATATTAGGCGGAAATGGGTACATCAATGACTATCCTACTGGTAGGATTTTGAGAGATGCAAAGCTGTATGAAATAGGTGCAGGAACTTCAGAAGTGAGGAGAATGTTAATTGGAAGAGCTTTAAATAATGAATACAAGTGA
Protein
MVVRLCVRRLLRKTANKTGHRCISHYPIDDHVFGLSEEQQQLRKMVFDFAQKELAPKAAEIDKENNFKELRPFWKKLGDLGLLGITASSDYGGTGGKYSDHCVIMEELSRASGGIALSYGAHSNLCVNQINRNGTEEQKSKYLPKLCSGEHIGALAMSEPGSGSDVVSMKLRAEKKGDYYVLNGNKFWITNGPDADVLVVYAKTNWSTSKQQHGISAFLIEKDYPGFSTAQKLDKLGMRGSNTGELVFEDCKVPAANLLGQENKGVYVLMSGLDLERLVLAAGPVGLMQAAIDTAFLYAHTRKQFGKNIGEFQLIQGKMADMYTTLSACRSYLYNVAKACDNGHVNSKDCAGVILYCAEKATQVALDAIQILGGNGYINDYPTGRILRDAKLYEIGAGTSEVRRMLIGRALNNEYK
Summary
Cofactor
FAD
Similarity
Belongs to the acyl-CoA dehydrogenase family.
Uniprot
B5UB85
D2SNL6
A0A2H1WEI4
A0A0N1ICI2
A0A212EZL0
A0A194QDM0
+ More
A0A2A4J6L8 E0W349 A0A0P4W309 E9GS77 A0A154P1H5 A0A0N7ZIY7 K7IUR0 A0A1S4F3T4 B0X633 U5EUB2 Q17GD3 A0A182YJH3 A0A182FFH5 A0A1Q3F1W5 A0A084WMZ7 A0A0P6EKI7 A0A182WI57 A0A182T4W4 A0A1B6KKR0 A0A336K0R5 A0A182INW0 A0A1B6FE49 A0A182H9F3 A0A232F5Q6 A0A182MS73 A0A2M4BNU3 A0A2M4A812 A0A2M4BNY6 W5JBT8 A0A182N7I3 T1E9J3 A0A0P6B5X2 A0A182UKR0 A0A182X250 A0A182L429 Q7QIC5 A0A182R9B6 A0A182PEX0 B3M691 A0A182V0F8 A0A2M3ZHG1 A0A1W4UEZ7 B3NBR7 A0A182I6B1 A0A2M3ZH71 Q29CZ7 B4H9H6 B4KZ54 B4K041 B4LEK2 A0A3M7RYJ7 B4HJZ3 A0A0A1XKN1 B4PG06 A0A3B0J6Q1 B4QM45 Q9VSL9 A0A0K8WEC0 A0A1I8NSK4 A0A034VY15 A0A034VTX1 A0A310SC54 D3TLC7 A0A023ESZ9 A0A1A9UJD5 A0A0L0BYS5 A0A1A9Z2Q7 A0A0M4EK03 B4MXU5 B7P6T5 A0A1B0BA97 A0A1A9YN39 A0A1L8DYZ7 A0A0K8RJD6 T1PCM6 A0A1I8MJ66 A0A1B0CM01 W8BYD7 D6WJK5 D3PGR5 C1C1U2 A0A2P2HZ63 A0A026WU13 A0A3L8DG85 A0A1W7RAA9 A0A1I8AG09 A0A1L8DYV7 A0A226EXE4 R7UHR6 A0A088AHV5 A0A087SWH9 A0A2A3EAW3 A0A067R9Z1 A0A2S2R6I3
A0A2A4J6L8 E0W349 A0A0P4W309 E9GS77 A0A154P1H5 A0A0N7ZIY7 K7IUR0 A0A1S4F3T4 B0X633 U5EUB2 Q17GD3 A0A182YJH3 A0A182FFH5 A0A1Q3F1W5 A0A084WMZ7 A0A0P6EKI7 A0A182WI57 A0A182T4W4 A0A1B6KKR0 A0A336K0R5 A0A182INW0 A0A1B6FE49 A0A182H9F3 A0A232F5Q6 A0A182MS73 A0A2M4BNU3 A0A2M4A812 A0A2M4BNY6 W5JBT8 A0A182N7I3 T1E9J3 A0A0P6B5X2 A0A182UKR0 A0A182X250 A0A182L429 Q7QIC5 A0A182R9B6 A0A182PEX0 B3M691 A0A182V0F8 A0A2M3ZHG1 A0A1W4UEZ7 B3NBR7 A0A182I6B1 A0A2M3ZH71 Q29CZ7 B4H9H6 B4KZ54 B4K041 B4LEK2 A0A3M7RYJ7 B4HJZ3 A0A0A1XKN1 B4PG06 A0A3B0J6Q1 B4QM45 Q9VSL9 A0A0K8WEC0 A0A1I8NSK4 A0A034VY15 A0A034VTX1 A0A310SC54 D3TLC7 A0A023ESZ9 A0A1A9UJD5 A0A0L0BYS5 A0A1A9Z2Q7 A0A0M4EK03 B4MXU5 B7P6T5 A0A1B0BA97 A0A1A9YN39 A0A1L8DYZ7 A0A0K8RJD6 T1PCM6 A0A1I8MJ66 A0A1B0CM01 W8BYD7 D6WJK5 D3PGR5 C1C1U2 A0A2P2HZ63 A0A026WU13 A0A3L8DG85 A0A1W7RAA9 A0A1I8AG09 A0A1L8DYV7 A0A226EXE4 R7UHR6 A0A088AHV5 A0A087SWH9 A0A2A3EAW3 A0A067R9Z1 A0A2S2R6I3
Pubmed
19121390
21040472
20074338
26354079
22118469
20566863
+ More
21292972 20075255 17510324 25244985 24438588 26483478 28648823 20920257 23761445 20966253 12364791 14747013 17210077 17994087 15632085 30375419 25830018 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 20353571 24945155 26108605 25315136 24495485 18362917 19820115 24508170 30249741 26392177 23254933 24845553
21292972 20075255 17510324 25244985 24438588 26483478 28648823 20920257 23761445 20966253 12364791 14747013 17210077 17994087 15632085 30375419 25830018 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 20353571 24945155 26108605 25315136 24495485 18362917 19820115 24508170 30249741 26392177 23254933 24845553
EMBL
BABH01013466
BABH01013467
AB458683
AB462483
BAG72196.1
BAG74561.1
+ More
EZ407145 ACX53707.1 ODYU01008136 SOQ51488.1 KQ460968 KPJ10212.1 AGBW02011262 OWR46923.1 KQ459324 KPJ01556.1 NWSH01003026 PCG67090.1 DS235882 EEB20055.1 GDRN01076512 JAI62883.1 GL732561 EFX77663.1 KQ434786 KZC05088.1 GDIP01240841 GDIQ01006328 LRGB01000781 JAI82560.1 JAN88409.1 KZS15972.1 AAZX01002655 DS232400 EDS41164.1 GANO01002402 JAB57469.1 CH477263 EAT45617.1 GFDL01013557 JAV21488.1 ATLV01024541 ATLV01024542 KE525352 KFB51591.1 GDIQ01061266 JAN33471.1 GEBQ01028223 GEBQ01021061 JAT11754.1 JAT18916.1 UFQS01000015 UFQT01000015 SSW97372.1 SSX17758.1 GECZ01021280 JAS48489.1 JXUM01030163 JXUM01030164 KQ560874 KXJ80523.1 NNAY01000902 OXU26005.1 AXCM01000817 GGFJ01005541 MBW54682.1 GGFK01003560 MBW36881.1 GGFJ01005542 MBW54683.1 ADMH02001849 ETN60883.1 GAMD01001297 JAB00294.1 GDIP01019394 JAM84321.1 AAAB01008807 EAA04728.3 CH902618 EDV39711.1 GGFM01007139 MBW27890.1 CH954178 EDV50663.1 APCN01003602 GGFM01007121 MBW27872.1 CH379070 EAL30617.1 CH479229 EDW36452.1 CH933809 EDW17851.1 CH916386 EDV94289.1 CH940647 EDW69087.1 REGN01002396 RNA28397.1 CH480815 EDW40729.1 GBXI01002785 JAD11507.1 CM000159 EDW93150.1 OUUW01000002 SPP77405.1 CM000363 CM002912 EDX09728.1 KMY98412.1 AE014296 AY089640 AAF50398.1 AAL90378.1 GDHF01002808 JAI49506.1 GAKP01012212 JAC46740.1 GAKP01012211 JAC46741.1 KQ794714 OAD47022.1 CCAG010009360 EZ422229 ADD18505.1 GAPW01001667 JAC11931.1 JRES01001151 KNC25141.1 CP012525 ALC43460.1 CH963876 EDW76864.1 ABJB010000880 ABJB010035133 ABJB010059759 ABJB010215077 DS647752 EEC02307.1 JXJN01010876 GFDF01002437 JAV11647.1 GADI01002800 JAA71008.1 KA646439 AFP61068.1 AJWK01017981 AJWK01017982 GAMC01008244 JAB98311.1 KQ971342 EFA04754.2 BT120821 ADD24461.1 BT080821 ACO15245.1 IACF01001202 LAB66916.1 KK107109 EZA59106.1 QOIP01000008 RLU19455.1 GFAH01000307 JAV48082.1 GFDF01002475 JAV11609.1 LNIX01000001 OXA61848.1 AMQN01008687 KB303737 ELU02822.1 KK112265 KFM57218.1 KZ288310 PBC28614.1 KK852601 KDR20512.1 GGMS01016418 MBY85621.1
EZ407145 ACX53707.1 ODYU01008136 SOQ51488.1 KQ460968 KPJ10212.1 AGBW02011262 OWR46923.1 KQ459324 KPJ01556.1 NWSH01003026 PCG67090.1 DS235882 EEB20055.1 GDRN01076512 JAI62883.1 GL732561 EFX77663.1 KQ434786 KZC05088.1 GDIP01240841 GDIQ01006328 LRGB01000781 JAI82560.1 JAN88409.1 KZS15972.1 AAZX01002655 DS232400 EDS41164.1 GANO01002402 JAB57469.1 CH477263 EAT45617.1 GFDL01013557 JAV21488.1 ATLV01024541 ATLV01024542 KE525352 KFB51591.1 GDIQ01061266 JAN33471.1 GEBQ01028223 GEBQ01021061 JAT11754.1 JAT18916.1 UFQS01000015 UFQT01000015 SSW97372.1 SSX17758.1 GECZ01021280 JAS48489.1 JXUM01030163 JXUM01030164 KQ560874 KXJ80523.1 NNAY01000902 OXU26005.1 AXCM01000817 GGFJ01005541 MBW54682.1 GGFK01003560 MBW36881.1 GGFJ01005542 MBW54683.1 ADMH02001849 ETN60883.1 GAMD01001297 JAB00294.1 GDIP01019394 JAM84321.1 AAAB01008807 EAA04728.3 CH902618 EDV39711.1 GGFM01007139 MBW27890.1 CH954178 EDV50663.1 APCN01003602 GGFM01007121 MBW27872.1 CH379070 EAL30617.1 CH479229 EDW36452.1 CH933809 EDW17851.1 CH916386 EDV94289.1 CH940647 EDW69087.1 REGN01002396 RNA28397.1 CH480815 EDW40729.1 GBXI01002785 JAD11507.1 CM000159 EDW93150.1 OUUW01000002 SPP77405.1 CM000363 CM002912 EDX09728.1 KMY98412.1 AE014296 AY089640 AAF50398.1 AAL90378.1 GDHF01002808 JAI49506.1 GAKP01012212 JAC46740.1 GAKP01012211 JAC46741.1 KQ794714 OAD47022.1 CCAG010009360 EZ422229 ADD18505.1 GAPW01001667 JAC11931.1 JRES01001151 KNC25141.1 CP012525 ALC43460.1 CH963876 EDW76864.1 ABJB010000880 ABJB010035133 ABJB010059759 ABJB010215077 DS647752 EEC02307.1 JXJN01010876 GFDF01002437 JAV11647.1 GADI01002800 JAA71008.1 KA646439 AFP61068.1 AJWK01017981 AJWK01017982 GAMC01008244 JAB98311.1 KQ971342 EFA04754.2 BT120821 ADD24461.1 BT080821 ACO15245.1 IACF01001202 LAB66916.1 KK107109 EZA59106.1 QOIP01000008 RLU19455.1 GFAH01000307 JAV48082.1 GFDF01002475 JAV11609.1 LNIX01000001 OXA61848.1 AMQN01008687 KB303737 ELU02822.1 KK112265 KFM57218.1 KZ288310 PBC28614.1 KK852601 KDR20512.1 GGMS01016418 MBY85621.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000218220
UP000009046
+ More
UP000000305 UP000076502 UP000076858 UP000002358 UP000002320 UP000008820 UP000076408 UP000069272 UP000030765 UP000075920 UP000075901 UP000075880 UP000069940 UP000249989 UP000215335 UP000075883 UP000000673 UP000075884 UP000075902 UP000076407 UP000075882 UP000007062 UP000075900 UP000075885 UP000007801 UP000075903 UP000192221 UP000008711 UP000075840 UP000001819 UP000008744 UP000009192 UP000001070 UP000008792 UP000276133 UP000001292 UP000002282 UP000268350 UP000000304 UP000000803 UP000095300 UP000092444 UP000078200 UP000037069 UP000092445 UP000092553 UP000007798 UP000001555 UP000092460 UP000092443 UP000095301 UP000092461 UP000007266 UP000053097 UP000279307 UP000095287 UP000198287 UP000014760 UP000005203 UP000054359 UP000242457 UP000027135
UP000000305 UP000076502 UP000076858 UP000002358 UP000002320 UP000008820 UP000076408 UP000069272 UP000030765 UP000075920 UP000075901 UP000075880 UP000069940 UP000249989 UP000215335 UP000075883 UP000000673 UP000075884 UP000075902 UP000076407 UP000075882 UP000007062 UP000075900 UP000075885 UP000007801 UP000075903 UP000192221 UP000008711 UP000075840 UP000001819 UP000008744 UP000009192 UP000001070 UP000008792 UP000276133 UP000001292 UP000002282 UP000268350 UP000000304 UP000000803 UP000095300 UP000092444 UP000078200 UP000037069 UP000092445 UP000092553 UP000007798 UP000001555 UP000092460 UP000092443 UP000095301 UP000092461 UP000007266 UP000053097 UP000279307 UP000095287 UP000198287 UP000014760 UP000005203 UP000054359 UP000242457 UP000027135
Interpro
IPR009100
AcylCoA_DH/oxidase_NM_dom
+ More
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR009075 AcylCo_DH/oxidase_C
IPR006089 Acyl-CoA_DH_CS
IPR013786 AcylCoA_DH/ox_N
IPR036250 AcylCo_DH-like_C
IPR037069 AcylCoA_DH/ox_N_sf
IPR034183 IVD
IPR001353 Proteasome_sua/b
IPR023333 Proteasome_suB-type
IPR029055 Ntn_hydrolases_N
IPR016050 Proteasome_bsu_CS
IPR000243 Pept_T1A_subB
IPR006091 Acyl-CoA_Oxase/DH_cen-dom
IPR009075 AcylCo_DH/oxidase_C
IPR006089 Acyl-CoA_DH_CS
IPR013786 AcylCoA_DH/ox_N
IPR036250 AcylCo_DH-like_C
IPR037069 AcylCoA_DH/ox_N_sf
IPR034183 IVD
IPR001353 Proteasome_sua/b
IPR023333 Proteasome_suB-type
IPR029055 Ntn_hydrolases_N
IPR016050 Proteasome_bsu_CS
IPR000243 Pept_T1A_subB
Gene 3D
CDD
ProteinModelPortal
B5UB85
D2SNL6
A0A2H1WEI4
A0A0N1ICI2
A0A212EZL0
A0A194QDM0
+ More
A0A2A4J6L8 E0W349 A0A0P4W309 E9GS77 A0A154P1H5 A0A0N7ZIY7 K7IUR0 A0A1S4F3T4 B0X633 U5EUB2 Q17GD3 A0A182YJH3 A0A182FFH5 A0A1Q3F1W5 A0A084WMZ7 A0A0P6EKI7 A0A182WI57 A0A182T4W4 A0A1B6KKR0 A0A336K0R5 A0A182INW0 A0A1B6FE49 A0A182H9F3 A0A232F5Q6 A0A182MS73 A0A2M4BNU3 A0A2M4A812 A0A2M4BNY6 W5JBT8 A0A182N7I3 T1E9J3 A0A0P6B5X2 A0A182UKR0 A0A182X250 A0A182L429 Q7QIC5 A0A182R9B6 A0A182PEX0 B3M691 A0A182V0F8 A0A2M3ZHG1 A0A1W4UEZ7 B3NBR7 A0A182I6B1 A0A2M3ZH71 Q29CZ7 B4H9H6 B4KZ54 B4K041 B4LEK2 A0A3M7RYJ7 B4HJZ3 A0A0A1XKN1 B4PG06 A0A3B0J6Q1 B4QM45 Q9VSL9 A0A0K8WEC0 A0A1I8NSK4 A0A034VY15 A0A034VTX1 A0A310SC54 D3TLC7 A0A023ESZ9 A0A1A9UJD5 A0A0L0BYS5 A0A1A9Z2Q7 A0A0M4EK03 B4MXU5 B7P6T5 A0A1B0BA97 A0A1A9YN39 A0A1L8DYZ7 A0A0K8RJD6 T1PCM6 A0A1I8MJ66 A0A1B0CM01 W8BYD7 D6WJK5 D3PGR5 C1C1U2 A0A2P2HZ63 A0A026WU13 A0A3L8DG85 A0A1W7RAA9 A0A1I8AG09 A0A1L8DYV7 A0A226EXE4 R7UHR6 A0A088AHV5 A0A087SWH9 A0A2A3EAW3 A0A067R9Z1 A0A2S2R6I3
A0A2A4J6L8 E0W349 A0A0P4W309 E9GS77 A0A154P1H5 A0A0N7ZIY7 K7IUR0 A0A1S4F3T4 B0X633 U5EUB2 Q17GD3 A0A182YJH3 A0A182FFH5 A0A1Q3F1W5 A0A084WMZ7 A0A0P6EKI7 A0A182WI57 A0A182T4W4 A0A1B6KKR0 A0A336K0R5 A0A182INW0 A0A1B6FE49 A0A182H9F3 A0A232F5Q6 A0A182MS73 A0A2M4BNU3 A0A2M4A812 A0A2M4BNY6 W5JBT8 A0A182N7I3 T1E9J3 A0A0P6B5X2 A0A182UKR0 A0A182X250 A0A182L429 Q7QIC5 A0A182R9B6 A0A182PEX0 B3M691 A0A182V0F8 A0A2M3ZHG1 A0A1W4UEZ7 B3NBR7 A0A182I6B1 A0A2M3ZH71 Q29CZ7 B4H9H6 B4KZ54 B4K041 B4LEK2 A0A3M7RYJ7 B4HJZ3 A0A0A1XKN1 B4PG06 A0A3B0J6Q1 B4QM45 Q9VSL9 A0A0K8WEC0 A0A1I8NSK4 A0A034VY15 A0A034VTX1 A0A310SC54 D3TLC7 A0A023ESZ9 A0A1A9UJD5 A0A0L0BYS5 A0A1A9Z2Q7 A0A0M4EK03 B4MXU5 B7P6T5 A0A1B0BA97 A0A1A9YN39 A0A1L8DYZ7 A0A0K8RJD6 T1PCM6 A0A1I8MJ66 A0A1B0CM01 W8BYD7 D6WJK5 D3PGR5 C1C1U2 A0A2P2HZ63 A0A026WU13 A0A3L8DG85 A0A1W7RAA9 A0A1I8AG09 A0A1L8DYV7 A0A226EXE4 R7UHR6 A0A088AHV5 A0A087SWH9 A0A2A3EAW3 A0A067R9Z1 A0A2S2R6I3
PDB
1IVH
E-value=5.24624e-168,
Score=1517
Ontologies
PATHWAY
GO
Topology
Subcellular location
Nucleus
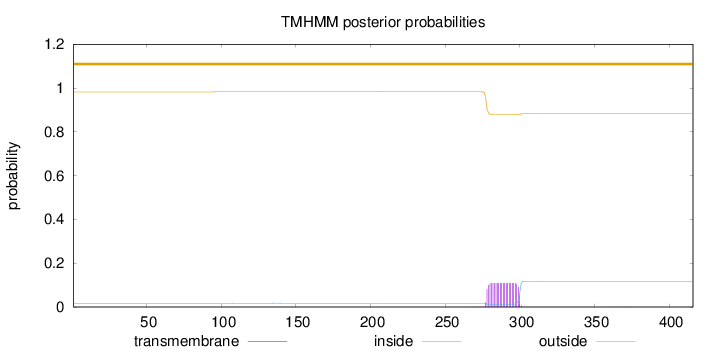
Length:
416
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.43588
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01754
outside
1 - 416
Population Genetic Test Statistics
Pi
245.37758
Theta
197.104815
Tajima's D
0.844714
CLR
0.194583
CSRT
0.615569221538923
Interpretation
Uncertain
