Gene
KWMTBOMO12946 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000235
Annotation
PREDICTED:_bifunctional_3'-phosphoadenosine_5'-phosphosulfate_synthase_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.275
Sequence
CDS
ATGGACGTCATCCGACTGCTGGAGTCCCAGGGTATCATTCCGAGCCTCGAGAGCAACCGTTCTGGAGTTGAAGAGCTCTTCATCCACGGGAACAGGCTCACCAGCGCCATCGAGGAGGCGGCCCGGCTACCACAGATTGAACTGACGACACTCGATTTGCAATGGGTTCAGGTTTTATCCGAAGGATGGGCGTATCCTTTAAAGGGATTTATGCGGGAAACCGAGTACTTACAGGCGCTTCATTCGAACTGCGTTACGCTGCCGGACGGGTCGCTAGTGAACCAGTCGGTGCCGGTGGTGTTGCCGGTGTCGGCCGAGCTGAAGGAGCGGCTGGCGGGCGCGGCGGCGGTGGCACTCACGCGCGGCGGCCGGCCCCTCGCCGTGCTGCGTGGCCCCGAGTTCTACCCGCACCGGAAGGAGGAGCGGTGCTCGCGGCAGTTCGGCATATACCACGCCGGTCACCCCTACATCAAGATGATCAAAGACTCCGGTGACTGGCTGGTAGGCGGCACCCTGGAGGTGTTCGAGCGGATCCGTTGGAACGACGGCCTGGACTCGTACCGCCTCACCCCCAACGAGCTCAGACAGAAGTTCAAGGACCTGGGGGCTGACGCCGTGTTCGCGTTCCAGTTGCGGAACCCGATCCACAACGGGCACGCGCTGCTGATGCAGGACACGCAGCGGCAACTGGTCGAGAGAGGATTCAAGAAACCCGTGCTCTTGTTACATCCGCTCGGCGGTTGGACGAAAGACGACGACGTGCCTCTAGAAGTGAGAATAAAACAACACAAAGCCGTGCTAAAGGAGGGAGTCTTGGATCCTAACTCCACTGTCCTGGCCATTTTCCCGTCCCCTATGATGTACGCCGGACCCACTGAGGTGCAGTGGCACGCCAAGTGTCGCATGAACGCCGGCGCCAACCACTACATAGTGGGGCGCGACCCGGCCGGCCTGCCGTACCCGCGCGGCGACGGCGACCTGTACGACCCGCGGCACGGCGCCATGCTGCTCAAGGTCGCGCCCGGACTCAACGACCTGGAGATAATTCCGTTCCGCGTGGCCGCCTACGATACGTCGGTGGGTAAGATGGCGTTCTTCGACCCGACGCGCAAAGAGGACTTCGACTTCATATCCGGAACGAGGATGAGAGGGCTCGCGAAGGCCGGCAAGGAGCCTCCGAGGGGCTTCATGGCGCCGACCGCCTGGAACGTTCTCGCCGAGTACTACCAGTCGATGAAATCCAAAATGGATACGAATTAA
Protein
MDVIRLLESQGIIPSLESNRSGVEELFIHGNRLTSAIEEAARLPQIELTTLDLQWVQVLSEGWAYPLKGFMRETEYLQALHSNCVTLPDGSLVNQSVPVVLPVSAELKERLAGAAAVALTRGGRPLAVLRGPEFYPHRKEERCSRQFGIYHAGHPYIKMIKDSGDWLVGGTLEVFERIRWNDGLDSYRLTPNELRQKFKDLGADAVFAFQLRNPIHNGHALLMQDTQRQLVERGFKKPVLLLHPLGGWTKDDDVPLEVRIKQHKAVLKEGVLDPNSTVLAIFPSPMMYAGPTEVQWHAKCRMNAGANHYIVGRDPAGLPYPRGDGDLYDPRHGAMLLKVAPGLNDLEIIPFRVAAYDTSVGKMAFFDPTRKEDFDFISGTRMRGLAKAGKEPPRGFMAPTAWNVLAEYYQSMKSKMDTN
Summary
Uniprot
A0A2A4IXA5
A0A2H1WEJ6
S4PXI3
A0A194Q7P1
A0A0N1PGA6
A0A2J7Q7Y1
+ More
A0A2J7Q7Y5 A0A182Q0A7 E0W351 A0A084W067 Q7PX77 F5HMV2 V5GYZ7 D7EKF1 A0A182UEY1 B7P3J8 A0A182L8N9 W8BGD4 A0A0T6B7E6 F5HMV3 A0A182X9G4 A0A182V205 A0A182MPX3 A0A1Y9IV51 A0A1Y9IVV1 A0A1Y9IVX0 A0A2M4BGK2 A0A182RA51 A0A0A1WK45 A0A182HKE9 A0A2M4BHH6 A0A2M4A1Z9 A0A2M4A1W1 Q179J5 A0A034W308 A0A2M4A240 A0A2M4A284 A0A0P4VVW0 A0A2M4A1W7 A0A182FRZ8 A0A0A9ZCS8 A0A0A9YY03 T1PKZ2 T1J7Q2 A0A034W5Q0 A0A2M3Z5Q2 A0A1B6JPF9 W5J5A6 A0A0K8S8J3 T1PL68 A0A1I8MDT4 A0A1S3J702 A0A023F5B2 A0A2M3ZH96 A0A1S3J7A6 A0A161MSV4 A0A224YLU0 A0A182NQP4 A0A131YPQ9 A0A131YG23 A0A1B6JTU7 A0A1S3J8K0 T1I8C2 G3MPV1 L7M6S3 A0A069DZC3 A0A023EUU6 A0A2M4CLM3 A0A131XHN2 A0A0K8TXW2 A0A182GHQ8 V5G3R4 A0A067RA86 A0A182JCQ3 A0A0L0CF95 A0A182YQ18 A0A1I8PAV5 A0A1E1XAX5 A0A1A9UQG6 A0A182PRS4 A0A1I8PAQ4 A0A2G8L145 A0A1I8PAN7 A0A1A9XDC9 D3TLJ1 A0A1B0GD56 A0A1Q3FZ31 A0A0K8TT79 V4BE20 A0A1Q3FYW9 A0A1Q3FYV1 A0A1B0C0A8 A0A1A9ZS86 K7J476 A0A0J7KS12 N6TNS0 B3DJF3 A0A3S0ZGJ2 K1R185 A0A2A3E6X3
A0A2J7Q7Y5 A0A182Q0A7 E0W351 A0A084W067 Q7PX77 F5HMV2 V5GYZ7 D7EKF1 A0A182UEY1 B7P3J8 A0A182L8N9 W8BGD4 A0A0T6B7E6 F5HMV3 A0A182X9G4 A0A182V205 A0A182MPX3 A0A1Y9IV51 A0A1Y9IVV1 A0A1Y9IVX0 A0A2M4BGK2 A0A182RA51 A0A0A1WK45 A0A182HKE9 A0A2M4BHH6 A0A2M4A1Z9 A0A2M4A1W1 Q179J5 A0A034W308 A0A2M4A240 A0A2M4A284 A0A0P4VVW0 A0A2M4A1W7 A0A182FRZ8 A0A0A9ZCS8 A0A0A9YY03 T1PKZ2 T1J7Q2 A0A034W5Q0 A0A2M3Z5Q2 A0A1B6JPF9 W5J5A6 A0A0K8S8J3 T1PL68 A0A1I8MDT4 A0A1S3J702 A0A023F5B2 A0A2M3ZH96 A0A1S3J7A6 A0A161MSV4 A0A224YLU0 A0A182NQP4 A0A131YPQ9 A0A131YG23 A0A1B6JTU7 A0A1S3J8K0 T1I8C2 G3MPV1 L7M6S3 A0A069DZC3 A0A023EUU6 A0A2M4CLM3 A0A131XHN2 A0A0K8TXW2 A0A182GHQ8 V5G3R4 A0A067RA86 A0A182JCQ3 A0A0L0CF95 A0A182YQ18 A0A1I8PAV5 A0A1E1XAX5 A0A1A9UQG6 A0A182PRS4 A0A1I8PAQ4 A0A2G8L145 A0A1I8PAN7 A0A1A9XDC9 D3TLJ1 A0A1B0GD56 A0A1Q3FZ31 A0A0K8TT79 V4BE20 A0A1Q3FYW9 A0A1Q3FYV1 A0A1B0C0A8 A0A1A9ZS86 K7J476 A0A0J7KS12 N6TNS0 B3DJF3 A0A3S0ZGJ2 K1R185 A0A2A3E6X3
Pubmed
23622113
26354079
20566863
24438588
12364791
14747013
+ More
17210077 25765539 18362917 19820115 20966253 24495485 25830018 17510324 25348373 27129103 25401762 26823975 20920257 23761445 25315136 25474469 28797301 26830274 22216098 25576852 26334808 24945155 28049606 26483478 24845553 26108605 25244985 28503490 29023486 20353571 26369729 23254933 20075255 23537049 23594743 22992520
17210077 25765539 18362917 19820115 20966253 24495485 25830018 17510324 25348373 27129103 25401762 26823975 20920257 23761445 25315136 25474469 28797301 26830274 22216098 25576852 26334808 24945155 28049606 26483478 24845553 26108605 25244985 28503490 29023486 20353571 26369729 23254933 20075255 23537049 23594743 22992520
EMBL
NWSH01005836
PCG63904.1
ODYU01008136
SOQ51489.1
GAIX01004988
JAA87572.1
+ More
KQ459324 KPJ01557.1 KQ460968 KPJ10213.1 NEVH01017000 PNF24694.1 PNF24692.1 AXCN02001509 DS235882 EEB20057.1 ATLV01019092 KE525262 KFB43611.1 AAAB01008987 EAA01759.4 EGK97625.1 GANP01008628 JAB75840.1 DS497744 EFA13134.1 ABJB010347738 ABJB010864233 DS629251 EEC01170.1 GAMC01014214 JAB92341.1 LJIG01009419 KRT83118.1 EGK97624.1 AXCM01000640 GGFJ01002930 MBW52071.1 GBXI01015412 JAC98879.1 APCN01000826 GGFJ01003351 MBW52492.1 GGFK01001458 MBW34779.1 GGFK01001483 MBW34804.1 CH477349 EAT42895.1 GAKP01009858 JAC49094.1 GGFK01001526 MBW34847.1 GGFK01001613 MBW34934.1 GDKW01001778 JAI54817.1 GGFK01001456 MBW34777.1 GBHO01009158 GBHO01009155 GBHO01001345 GDHC01015777 JAG34446.1 JAG34449.1 JAG42259.1 JAQ02852.1 GBHO01009156 GBHO01009154 GBHO01001285 GDHC01019910 GDHC01005794 JAG34448.1 JAG34450.1 JAG42319.1 JAP98718.1 JAQ12835.1 KA649384 AFP64013.1 JH431938 GAKP01009859 JAC49093.1 GGFM01003098 MBW23849.1 GECU01006560 JAT01147.1 ADMH02002101 ETN59166.1 GBRD01016229 GBRD01014861 GBRD01014860 GBRD01003037 JAG49597.1 KA649497 AFP64126.1 GBBI01002418 JAC16294.1 GGFM01007153 MBW27904.1 GEMB01000561 JAS02569.1 GFPF01005563 MAA16709.1 GEDV01007304 JAP81253.1 GEDV01011127 JAP77430.1 GECU01005065 JAT02642.1 ACPB03000011 JO843902 AEO35519.1 GACK01005038 JAA59996.1 GBGD01000890 JAC87999.1 GAPW01000388 JAC13210.1 GGFL01002005 MBW66183.1 GEFH01001992 JAP66589.1 GDHF01033015 JAI19299.1 JXUM01064346 JXUM01064347 KQ562293 KXJ76223.1 GALX01003801 JAB64665.1 KK852601 KDR20503.1 JRES01000484 KNC30915.1 GFAC01002801 JAT96387.1 MRZV01000268 PIK53977.1 EZ422293 ADD18569.1 CCAG010002318 GFDL01002313 JAV32732.1 GDAI01000245 JAI17358.1 KB199905 ESP04012.1 GFDL01002325 JAV32720.1 GFDL01002318 JAV32727.1 JXJN01023506 AAZX01003267 LBMM01003797 KMQ93123.1 APGK01057391 APGK01057392 KB741280 ENN70895.1 CR788312 BC163464 BC163465 AAI63464.1 RQTK01000608 RUS77120.1 JH817156 EKC37239.1 KZ288361 PBC26976.1
KQ459324 KPJ01557.1 KQ460968 KPJ10213.1 NEVH01017000 PNF24694.1 PNF24692.1 AXCN02001509 DS235882 EEB20057.1 ATLV01019092 KE525262 KFB43611.1 AAAB01008987 EAA01759.4 EGK97625.1 GANP01008628 JAB75840.1 DS497744 EFA13134.1 ABJB010347738 ABJB010864233 DS629251 EEC01170.1 GAMC01014214 JAB92341.1 LJIG01009419 KRT83118.1 EGK97624.1 AXCM01000640 GGFJ01002930 MBW52071.1 GBXI01015412 JAC98879.1 APCN01000826 GGFJ01003351 MBW52492.1 GGFK01001458 MBW34779.1 GGFK01001483 MBW34804.1 CH477349 EAT42895.1 GAKP01009858 JAC49094.1 GGFK01001526 MBW34847.1 GGFK01001613 MBW34934.1 GDKW01001778 JAI54817.1 GGFK01001456 MBW34777.1 GBHO01009158 GBHO01009155 GBHO01001345 GDHC01015777 JAG34446.1 JAG34449.1 JAG42259.1 JAQ02852.1 GBHO01009156 GBHO01009154 GBHO01001285 GDHC01019910 GDHC01005794 JAG34448.1 JAG34450.1 JAG42319.1 JAP98718.1 JAQ12835.1 KA649384 AFP64013.1 JH431938 GAKP01009859 JAC49093.1 GGFM01003098 MBW23849.1 GECU01006560 JAT01147.1 ADMH02002101 ETN59166.1 GBRD01016229 GBRD01014861 GBRD01014860 GBRD01003037 JAG49597.1 KA649497 AFP64126.1 GBBI01002418 JAC16294.1 GGFM01007153 MBW27904.1 GEMB01000561 JAS02569.1 GFPF01005563 MAA16709.1 GEDV01007304 JAP81253.1 GEDV01011127 JAP77430.1 GECU01005065 JAT02642.1 ACPB03000011 JO843902 AEO35519.1 GACK01005038 JAA59996.1 GBGD01000890 JAC87999.1 GAPW01000388 JAC13210.1 GGFL01002005 MBW66183.1 GEFH01001992 JAP66589.1 GDHF01033015 JAI19299.1 JXUM01064346 JXUM01064347 KQ562293 KXJ76223.1 GALX01003801 JAB64665.1 KK852601 KDR20503.1 JRES01000484 KNC30915.1 GFAC01002801 JAT96387.1 MRZV01000268 PIK53977.1 EZ422293 ADD18569.1 CCAG010002318 GFDL01002313 JAV32732.1 GDAI01000245 JAI17358.1 KB199905 ESP04012.1 GFDL01002325 JAV32720.1 GFDL01002318 JAV32727.1 JXJN01023506 AAZX01003267 LBMM01003797 KMQ93123.1 APGK01057391 APGK01057392 KB741280 ENN70895.1 CR788312 BC163464 BC163465 AAI63464.1 RQTK01000608 RUS77120.1 JH817156 EKC37239.1 KZ288361 PBC26976.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000235965
UP000075886
UP000009046
+ More
UP000030765 UP000007062 UP000007266 UP000075902 UP000001555 UP000075882 UP000076407 UP000075903 UP000075883 UP000075920 UP000075900 UP000075840 UP000008820 UP000069272 UP000000673 UP000095301 UP000085678 UP000075884 UP000015103 UP000069940 UP000249989 UP000027135 UP000075880 UP000037069 UP000076408 UP000095300 UP000078200 UP000075885 UP000230750 UP000092443 UP000092444 UP000030746 UP000092460 UP000092445 UP000002358 UP000036403 UP000019118 UP000000437 UP000271974 UP000005408 UP000242457
UP000030765 UP000007062 UP000007266 UP000075902 UP000001555 UP000075882 UP000076407 UP000075903 UP000075883 UP000075920 UP000075900 UP000075840 UP000008820 UP000069272 UP000000673 UP000095301 UP000085678 UP000075884 UP000015103 UP000069940 UP000249989 UP000027135 UP000075880 UP000037069 UP000076408 UP000095300 UP000078200 UP000075885 UP000230750 UP000092443 UP000092444 UP000030746 UP000092460 UP000092445 UP000002358 UP000036403 UP000019118 UP000000437 UP000271974 UP000005408 UP000242457
Interpro
IPR002891
APS_kinase
+ More
IPR027417 P-loop_NTPase
IPR025980 ATP-Sase_PUA-like_dom
IPR002650 Sulphate_adenylyltransferase
IPR014729 Rossmann-like_a/b/a_fold
IPR015947 PUA-like_sf
IPR024951 Sulfurylase_cat_dom
IPR016267 UDPGP_trans
IPR002618 UDPGP_fam
IPR029044 Nucleotide-diphossugar_trans
IPR012934 Znf_AD
IPR031632 SVIP
IPR027417 P-loop_NTPase
IPR025980 ATP-Sase_PUA-like_dom
IPR002650 Sulphate_adenylyltransferase
IPR014729 Rossmann-like_a/b/a_fold
IPR015947 PUA-like_sf
IPR024951 Sulfurylase_cat_dom
IPR016267 UDPGP_trans
IPR002618 UDPGP_fam
IPR029044 Nucleotide-diphossugar_trans
IPR012934 Znf_AD
IPR031632 SVIP
Gene 3D
ProteinModelPortal
A0A2A4IXA5
A0A2H1WEJ6
S4PXI3
A0A194Q7P1
A0A0N1PGA6
A0A2J7Q7Y1
+ More
A0A2J7Q7Y5 A0A182Q0A7 E0W351 A0A084W067 Q7PX77 F5HMV2 V5GYZ7 D7EKF1 A0A182UEY1 B7P3J8 A0A182L8N9 W8BGD4 A0A0T6B7E6 F5HMV3 A0A182X9G4 A0A182V205 A0A182MPX3 A0A1Y9IV51 A0A1Y9IVV1 A0A1Y9IVX0 A0A2M4BGK2 A0A182RA51 A0A0A1WK45 A0A182HKE9 A0A2M4BHH6 A0A2M4A1Z9 A0A2M4A1W1 Q179J5 A0A034W308 A0A2M4A240 A0A2M4A284 A0A0P4VVW0 A0A2M4A1W7 A0A182FRZ8 A0A0A9ZCS8 A0A0A9YY03 T1PKZ2 T1J7Q2 A0A034W5Q0 A0A2M3Z5Q2 A0A1B6JPF9 W5J5A6 A0A0K8S8J3 T1PL68 A0A1I8MDT4 A0A1S3J702 A0A023F5B2 A0A2M3ZH96 A0A1S3J7A6 A0A161MSV4 A0A224YLU0 A0A182NQP4 A0A131YPQ9 A0A131YG23 A0A1B6JTU7 A0A1S3J8K0 T1I8C2 G3MPV1 L7M6S3 A0A069DZC3 A0A023EUU6 A0A2M4CLM3 A0A131XHN2 A0A0K8TXW2 A0A182GHQ8 V5G3R4 A0A067RA86 A0A182JCQ3 A0A0L0CF95 A0A182YQ18 A0A1I8PAV5 A0A1E1XAX5 A0A1A9UQG6 A0A182PRS4 A0A1I8PAQ4 A0A2G8L145 A0A1I8PAN7 A0A1A9XDC9 D3TLJ1 A0A1B0GD56 A0A1Q3FZ31 A0A0K8TT79 V4BE20 A0A1Q3FYW9 A0A1Q3FYV1 A0A1B0C0A8 A0A1A9ZS86 K7J476 A0A0J7KS12 N6TNS0 B3DJF3 A0A3S0ZGJ2 K1R185 A0A2A3E6X3
A0A2J7Q7Y5 A0A182Q0A7 E0W351 A0A084W067 Q7PX77 F5HMV2 V5GYZ7 D7EKF1 A0A182UEY1 B7P3J8 A0A182L8N9 W8BGD4 A0A0T6B7E6 F5HMV3 A0A182X9G4 A0A182V205 A0A182MPX3 A0A1Y9IV51 A0A1Y9IVV1 A0A1Y9IVX0 A0A2M4BGK2 A0A182RA51 A0A0A1WK45 A0A182HKE9 A0A2M4BHH6 A0A2M4A1Z9 A0A2M4A1W1 Q179J5 A0A034W308 A0A2M4A240 A0A2M4A284 A0A0P4VVW0 A0A2M4A1W7 A0A182FRZ8 A0A0A9ZCS8 A0A0A9YY03 T1PKZ2 T1J7Q2 A0A034W5Q0 A0A2M3Z5Q2 A0A1B6JPF9 W5J5A6 A0A0K8S8J3 T1PL68 A0A1I8MDT4 A0A1S3J702 A0A023F5B2 A0A2M3ZH96 A0A1S3J7A6 A0A161MSV4 A0A224YLU0 A0A182NQP4 A0A131YPQ9 A0A131YG23 A0A1B6JTU7 A0A1S3J8K0 T1I8C2 G3MPV1 L7M6S3 A0A069DZC3 A0A023EUU6 A0A2M4CLM3 A0A131XHN2 A0A0K8TXW2 A0A182GHQ8 V5G3R4 A0A067RA86 A0A182JCQ3 A0A0L0CF95 A0A182YQ18 A0A1I8PAV5 A0A1E1XAX5 A0A1A9UQG6 A0A182PRS4 A0A1I8PAQ4 A0A2G8L145 A0A1I8PAN7 A0A1A9XDC9 D3TLJ1 A0A1B0GD56 A0A1Q3FZ31 A0A0K8TT79 V4BE20 A0A1Q3FYW9 A0A1Q3FYV1 A0A1B0C0A8 A0A1A9ZS86 K7J476 A0A0J7KS12 N6TNS0 B3DJF3 A0A3S0ZGJ2 K1R185 A0A2A3E6X3
PDB
1XNJ
E-value=1.96854e-158,
Score=1434
Ontologies
PATHWAY
GO
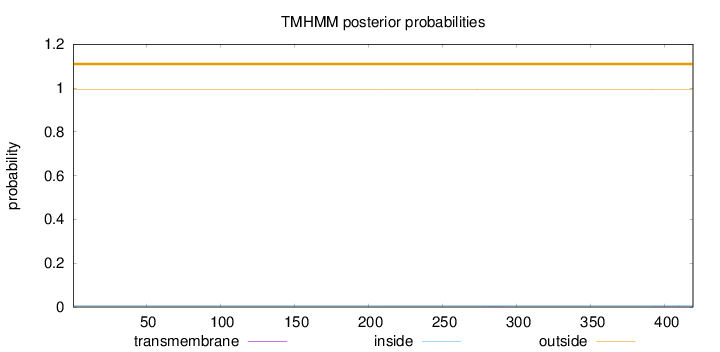
Topology
Length:
419
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01012
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00582
outside
1 - 419
Population Genetic Test Statistics
Pi
168.740702
Theta
175.784241
Tajima's D
0.318385
CLR
0.369926
CSRT
0.457977101144943
Interpretation
Possibly Positive selection
