Gene
KWMTBOMO12945
Pre Gene Modal
BGIBMGA000234
Annotation
adenylsulphate_kinase?_partial_[Papilio_polytes]
Full name
Adenylyl-sulfate kinase
Location in the cell
Mitochondrial Reliability : 1.081
Sequence
CDS
ATGGGTGGACGAGCTCACAGCCCACCTGGTGTTAAGTGGTTACTGGAGCCCATGGACATCTACAACTGCGCTCAGGTGGCCACGAATGTCGTCGAGCAGAAGCATCAAGTGTCCAGGGCCAAGAGGAGCAGAGCTCTTGGAAGCAGAGCGTTCAGGGGTAGCACCGTCTGGTTCACGGGGCTAAGCGGCGCTGGTAAAACCAGTATTGCCTTCGCTCTTGAAGCCTACTTGGTCTCCAAAGGAATCCCGGCTTACGGCCTCGATGGAGACAATATCAGGACTGGTCTGAACAAAAATCTTGGTTTCACGAAAGAAGATCGCGAGGAGAACATACGGCGTGTTGCCGAAGTTGCCAAGCTGTTTGCAGACAGCGGCGTCGTTTGTCTGTGCAGTTTTGTGTCGCCATTCGCTGAGGTTTTTCTGTGA
Protein
MGGRAHSPPGVKWLLEPMDIYNCAQVATNVVEQKHQVSRAKRSRALGSRAFRGSTVWFTGLSGAGKTSIAFALEAYLVSKGIPAYGLDGDNIRTGLNKNLGFTKEDREENIRRVAEVAKLFADSGVVCLCSFVSPFAEVFL
Summary
Description
Catalyzes the synthesis of activated sulfate.
Catalytic Activity
adenosine 5'-phosphosulfate + ATP = 3'-phosphoadenylyl sulfate + ADP + H(+)
Similarity
Belongs to the APS kinase family.
Uniprot
I4DR27
A0A2A4IXA5
S4PXI3
A0A194Q7P1
A0A0N1PGA6
A0A0A9ZCS8
+ More
A0A0A9YY03 A0A1B6LXE6 A0A1W4XSM0 T1I8C2 A0A1B6LSS0 A0A1B6KS59 V5G3R4 A0A2P8XV41 A0A2J7Q7Y5 A0A2J7Q7Y1 A0A067RA86 D7EKF1 A0A336LDL0 A0A336MY56 A0A0T6B7E6 A0A182T4K7 A0A0M4EHR4 A0A023F5B2 A0A069DZC3 A0A1A9X117 A0A1B6E2A6 A0A0P4VVW0 A0A1Y1N7K1 A0A1B6DDZ9 A0A1Y9IV51 Q7PX77 A0A2M3Z5Q2 E0W351 A0A1Y9IVV1 A0A182JS09 A0A2M3ZH96 A0A0Q9WXL2 A0A2M4CLM3 A0A2M4BHH6 A0A2M4A284 A0A1B6G0A5 A0A2M4A240 A0A224XMA7 A0A1B0Y0A8 F5HMV3 A0A1B6JPF9 A0A0Q9WVX5 A0A1B6ER44 B4L0X0 B4LBM2 A0A182V205 A0A0Q9XDE4 A0A182PRS4 A0A182YQ18 F5HMV2 A0A1Y9IVX0 W5J5A6 A0A182JCQ3 A0A182HKE9 A0A182UEY1 A0A182FRZ8 A0A182X9G4 A0A182L8N9 A0A2M4A1W7 A0A0Q9XDC0 A0A2M4A1Z9 A0A2M4A1W1 A0A2M4BGK2 A0A084W067 A0A182GHQ8 A0A023EUU6 A0A0Q9WLZ1 A0A182MPX3 B4QRC1 A0A0J9RYA0 B3M8T9 A0A1J1IIW1 A0A0R1DZH2 A0A0P9C5D7 A0A0J9UPH8 A0A1J1IML3 Q961A8 B4IXP3 B3NDZ8 A0A1B0CZQ5 B4PFX1 A0A1Q3FYW9 Q7KUT8 Q9VW48 B0WFJ9 A0A0Q5UC99 A0A182Q0A7 Q179J5 A0A0L0CF95 A0A1B0GD56 T1PKZ2 A0A1Q3FYV1 A0A182NQP4 A0A0P8ZWA1
A0A0A9YY03 A0A1B6LXE6 A0A1W4XSM0 T1I8C2 A0A1B6LSS0 A0A1B6KS59 V5G3R4 A0A2P8XV41 A0A2J7Q7Y5 A0A2J7Q7Y1 A0A067RA86 D7EKF1 A0A336LDL0 A0A336MY56 A0A0T6B7E6 A0A182T4K7 A0A0M4EHR4 A0A023F5B2 A0A069DZC3 A0A1A9X117 A0A1B6E2A6 A0A0P4VVW0 A0A1Y1N7K1 A0A1B6DDZ9 A0A1Y9IV51 Q7PX77 A0A2M3Z5Q2 E0W351 A0A1Y9IVV1 A0A182JS09 A0A2M3ZH96 A0A0Q9WXL2 A0A2M4CLM3 A0A2M4BHH6 A0A2M4A284 A0A1B6G0A5 A0A2M4A240 A0A224XMA7 A0A1B0Y0A8 F5HMV3 A0A1B6JPF9 A0A0Q9WVX5 A0A1B6ER44 B4L0X0 B4LBM2 A0A182V205 A0A0Q9XDE4 A0A182PRS4 A0A182YQ18 F5HMV2 A0A1Y9IVX0 W5J5A6 A0A182JCQ3 A0A182HKE9 A0A182UEY1 A0A182FRZ8 A0A182X9G4 A0A182L8N9 A0A2M4A1W7 A0A0Q9XDC0 A0A2M4A1Z9 A0A2M4A1W1 A0A2M4BGK2 A0A084W067 A0A182GHQ8 A0A023EUU6 A0A0Q9WLZ1 A0A182MPX3 B4QRC1 A0A0J9RYA0 B3M8T9 A0A1J1IIW1 A0A0R1DZH2 A0A0P9C5D7 A0A0J9UPH8 A0A1J1IML3 Q961A8 B4IXP3 B3NDZ8 A0A1B0CZQ5 B4PFX1 A0A1Q3FYW9 Q7KUT8 Q9VW48 B0WFJ9 A0A0Q5UC99 A0A182Q0A7 Q179J5 A0A0L0CF95 A0A1B0GD56 T1PKZ2 A0A1Q3FYV1 A0A182NQP4 A0A0P8ZWA1
EC Number
2.7.1.25
Pubmed
22651552
23622113
26354079
25401762
26823975
29403074
+ More
24845553 18362917 19820115 25474469 26334808 27129103 28004739 12364791 14747013 17210077 20566863 17994087 25244985 20920257 23761445 20966253 24438588 26483478 24945155 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9431815 17510324 26108605
24845553 18362917 19820115 25474469 26334808 27129103 28004739 12364791 14747013 17210077 20566863 17994087 25244985 20920257 23761445 20966253 24438588 26483478 24945155 22936249 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 9431815 17510324 26108605
EMBL
AK404910
BAM20367.1
NWSH01005836
PCG63904.1
GAIX01004988
JAA87572.1
+ More
KQ459324 KPJ01557.1 KQ460968 KPJ10213.1 GBHO01009158 GBHO01009155 GBHO01001345 GDHC01015777 JAG34446.1 JAG34449.1 JAG42259.1 JAQ02852.1 GBHO01009156 GBHO01009154 GBHO01001285 GDHC01019910 GDHC01005794 JAG34448.1 JAG34450.1 JAG42319.1 JAP98718.1 JAQ12835.1 GEBQ01011601 JAT28376.1 ACPB03000011 GEBQ01013242 JAT26735.1 GEBQ01025698 JAT14279.1 GALX01003801 JAB64665.1 PYGN01001300 PSN35865.1 NEVH01017000 PNF24692.1 PNF24694.1 KK852601 KDR20503.1 DS497744 EFA13134.1 UFQS01003803 UFQT01003803 SSX15946.1 SSX35284.1 UFQS01003881 UFQT01003881 SSX16009.1 SSX35342.1 LJIG01009419 KRT83118.1 CP012525 ALC44165.1 GBBI01002418 JAC16294.1 GBGD01000890 JAC87999.1 GEDC01005230 JAS32068.1 GDKW01001778 JAI54817.1 GEZM01015498 JAV91597.1 GEDC01013384 JAS23914.1 AAAB01008987 EAA01759.4 GGFM01003098 MBW23849.1 DS235882 EEB20057.1 GGFM01007153 MBW27904.1 CH940647 KRF85141.1 GGFL01002005 MBW66183.1 GGFJ01003351 MBW52492.1 GGFK01001613 MBW34934.1 GECZ01013892 JAS55877.1 GGFK01001526 MBW34847.1 GFTR01007275 JAW09151.1 KT000398 ANI85996.1 EGK97624.1 GECU01006560 JAT01147.1 KRF85140.1 GECZ01029556 JAS40213.1 CH933809 EDW19220.1 EDW70832.1 KRG06556.1 EGK97625.1 ADMH02002101 ETN59166.1 APCN01000826 GGFK01001456 MBW34777.1 KRG06555.1 GGFK01001458 MBW34779.1 GGFK01001483 MBW34804.1 GGFJ01002930 MBW52071.1 ATLV01019092 KE525262 KFB43611.1 JXUM01064346 JXUM01064347 KQ562293 KXJ76223.1 GAPW01000388 JAC13210.1 KRF85142.1 AXCM01000640 CM000363 EDX11134.1 CM002912 KMZ00627.1 CH902618 EDV41090.1 CVRI01000052 CRK99706.1 CM000159 KRK02341.1 KPU78919.1 KMZ00626.1 CRK99705.1 AY051724 AE014296 AAK93148.1 AAN11636.1 AAN11637.1 AAN11638.1 AGB94765.1 CH916366 EDV97505.1 CH954178 EDV52422.1 AJVK01020938 AJVK01020939 EDW95268.1 GFDL01002325 JAV32720.1 BT100002 AAF49102.2 ABI31264.1 ACX54886.1 AGB94766.1 Y12861 CAA73368.1 DS231918 EDS26267.1 KQS44269.1 AXCN02001509 CH477349 EAT42895.1 JRES01000484 KNC30915.1 CCAG010002318 KA649384 AFP64013.1 GFDL01002318 JAV32727.1 KPU78920.1
KQ459324 KPJ01557.1 KQ460968 KPJ10213.1 GBHO01009158 GBHO01009155 GBHO01001345 GDHC01015777 JAG34446.1 JAG34449.1 JAG42259.1 JAQ02852.1 GBHO01009156 GBHO01009154 GBHO01001285 GDHC01019910 GDHC01005794 JAG34448.1 JAG34450.1 JAG42319.1 JAP98718.1 JAQ12835.1 GEBQ01011601 JAT28376.1 ACPB03000011 GEBQ01013242 JAT26735.1 GEBQ01025698 JAT14279.1 GALX01003801 JAB64665.1 PYGN01001300 PSN35865.1 NEVH01017000 PNF24692.1 PNF24694.1 KK852601 KDR20503.1 DS497744 EFA13134.1 UFQS01003803 UFQT01003803 SSX15946.1 SSX35284.1 UFQS01003881 UFQT01003881 SSX16009.1 SSX35342.1 LJIG01009419 KRT83118.1 CP012525 ALC44165.1 GBBI01002418 JAC16294.1 GBGD01000890 JAC87999.1 GEDC01005230 JAS32068.1 GDKW01001778 JAI54817.1 GEZM01015498 JAV91597.1 GEDC01013384 JAS23914.1 AAAB01008987 EAA01759.4 GGFM01003098 MBW23849.1 DS235882 EEB20057.1 GGFM01007153 MBW27904.1 CH940647 KRF85141.1 GGFL01002005 MBW66183.1 GGFJ01003351 MBW52492.1 GGFK01001613 MBW34934.1 GECZ01013892 JAS55877.1 GGFK01001526 MBW34847.1 GFTR01007275 JAW09151.1 KT000398 ANI85996.1 EGK97624.1 GECU01006560 JAT01147.1 KRF85140.1 GECZ01029556 JAS40213.1 CH933809 EDW19220.1 EDW70832.1 KRG06556.1 EGK97625.1 ADMH02002101 ETN59166.1 APCN01000826 GGFK01001456 MBW34777.1 KRG06555.1 GGFK01001458 MBW34779.1 GGFK01001483 MBW34804.1 GGFJ01002930 MBW52071.1 ATLV01019092 KE525262 KFB43611.1 JXUM01064346 JXUM01064347 KQ562293 KXJ76223.1 GAPW01000388 JAC13210.1 KRF85142.1 AXCM01000640 CM000363 EDX11134.1 CM002912 KMZ00627.1 CH902618 EDV41090.1 CVRI01000052 CRK99706.1 CM000159 KRK02341.1 KPU78919.1 KMZ00626.1 CRK99705.1 AY051724 AE014296 AAK93148.1 AAN11636.1 AAN11637.1 AAN11638.1 AGB94765.1 CH916366 EDV97505.1 CH954178 EDV52422.1 AJVK01020938 AJVK01020939 EDW95268.1 GFDL01002325 JAV32720.1 BT100002 AAF49102.2 ABI31264.1 ACX54886.1 AGB94766.1 Y12861 CAA73368.1 DS231918 EDS26267.1 KQS44269.1 AXCN02001509 CH477349 EAT42895.1 JRES01000484 KNC30915.1 CCAG010002318 KA649384 AFP64013.1 GFDL01002318 JAV32727.1 KPU78920.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000192223
UP000015103
UP000245037
+ More
UP000235965 UP000027135 UP000007266 UP000075901 UP000092553 UP000091820 UP000075920 UP000007062 UP000009046 UP000075881 UP000008792 UP000009192 UP000075903 UP000075885 UP000076408 UP000000673 UP000075880 UP000075840 UP000075902 UP000069272 UP000076407 UP000075882 UP000030765 UP000069940 UP000249989 UP000075883 UP000000304 UP000007801 UP000183832 UP000002282 UP000000803 UP000001070 UP000008711 UP000092462 UP000002320 UP000075886 UP000008820 UP000037069 UP000092444 UP000075884
UP000235965 UP000027135 UP000007266 UP000075901 UP000092553 UP000091820 UP000075920 UP000007062 UP000009046 UP000075881 UP000008792 UP000009192 UP000075903 UP000075885 UP000076408 UP000000673 UP000075880 UP000075840 UP000075902 UP000069272 UP000076407 UP000075882 UP000030765 UP000069940 UP000249989 UP000075883 UP000000304 UP000007801 UP000183832 UP000002282 UP000000803 UP000001070 UP000008711 UP000092462 UP000002320 UP000075886 UP000008820 UP000037069 UP000092444 UP000075884
Interpro
Gene 3D
ProteinModelPortal
I4DR27
A0A2A4IXA5
S4PXI3
A0A194Q7P1
A0A0N1PGA6
A0A0A9ZCS8
+ More
A0A0A9YY03 A0A1B6LXE6 A0A1W4XSM0 T1I8C2 A0A1B6LSS0 A0A1B6KS59 V5G3R4 A0A2P8XV41 A0A2J7Q7Y5 A0A2J7Q7Y1 A0A067RA86 D7EKF1 A0A336LDL0 A0A336MY56 A0A0T6B7E6 A0A182T4K7 A0A0M4EHR4 A0A023F5B2 A0A069DZC3 A0A1A9X117 A0A1B6E2A6 A0A0P4VVW0 A0A1Y1N7K1 A0A1B6DDZ9 A0A1Y9IV51 Q7PX77 A0A2M3Z5Q2 E0W351 A0A1Y9IVV1 A0A182JS09 A0A2M3ZH96 A0A0Q9WXL2 A0A2M4CLM3 A0A2M4BHH6 A0A2M4A284 A0A1B6G0A5 A0A2M4A240 A0A224XMA7 A0A1B0Y0A8 F5HMV3 A0A1B6JPF9 A0A0Q9WVX5 A0A1B6ER44 B4L0X0 B4LBM2 A0A182V205 A0A0Q9XDE4 A0A182PRS4 A0A182YQ18 F5HMV2 A0A1Y9IVX0 W5J5A6 A0A182JCQ3 A0A182HKE9 A0A182UEY1 A0A182FRZ8 A0A182X9G4 A0A182L8N9 A0A2M4A1W7 A0A0Q9XDC0 A0A2M4A1Z9 A0A2M4A1W1 A0A2M4BGK2 A0A084W067 A0A182GHQ8 A0A023EUU6 A0A0Q9WLZ1 A0A182MPX3 B4QRC1 A0A0J9RYA0 B3M8T9 A0A1J1IIW1 A0A0R1DZH2 A0A0P9C5D7 A0A0J9UPH8 A0A1J1IML3 Q961A8 B4IXP3 B3NDZ8 A0A1B0CZQ5 B4PFX1 A0A1Q3FYW9 Q7KUT8 Q9VW48 B0WFJ9 A0A0Q5UC99 A0A182Q0A7 Q179J5 A0A0L0CF95 A0A1B0GD56 T1PKZ2 A0A1Q3FYV1 A0A182NQP4 A0A0P8ZWA1
A0A0A9YY03 A0A1B6LXE6 A0A1W4XSM0 T1I8C2 A0A1B6LSS0 A0A1B6KS59 V5G3R4 A0A2P8XV41 A0A2J7Q7Y5 A0A2J7Q7Y1 A0A067RA86 D7EKF1 A0A336LDL0 A0A336MY56 A0A0T6B7E6 A0A182T4K7 A0A0M4EHR4 A0A023F5B2 A0A069DZC3 A0A1A9X117 A0A1B6E2A6 A0A0P4VVW0 A0A1Y1N7K1 A0A1B6DDZ9 A0A1Y9IV51 Q7PX77 A0A2M3Z5Q2 E0W351 A0A1Y9IVV1 A0A182JS09 A0A2M3ZH96 A0A0Q9WXL2 A0A2M4CLM3 A0A2M4BHH6 A0A2M4A284 A0A1B6G0A5 A0A2M4A240 A0A224XMA7 A0A1B0Y0A8 F5HMV3 A0A1B6JPF9 A0A0Q9WVX5 A0A1B6ER44 B4L0X0 B4LBM2 A0A182V205 A0A0Q9XDE4 A0A182PRS4 A0A182YQ18 F5HMV2 A0A1Y9IVX0 W5J5A6 A0A182JCQ3 A0A182HKE9 A0A182UEY1 A0A182FRZ8 A0A182X9G4 A0A182L8N9 A0A2M4A1W7 A0A0Q9XDC0 A0A2M4A1Z9 A0A2M4A1W1 A0A2M4BGK2 A0A084W067 A0A182GHQ8 A0A023EUU6 A0A0Q9WLZ1 A0A182MPX3 B4QRC1 A0A0J9RYA0 B3M8T9 A0A1J1IIW1 A0A0R1DZH2 A0A0P9C5D7 A0A0J9UPH8 A0A1J1IML3 Q961A8 B4IXP3 B3NDZ8 A0A1B0CZQ5 B4PFX1 A0A1Q3FYW9 Q7KUT8 Q9VW48 B0WFJ9 A0A0Q5UC99 A0A182Q0A7 Q179J5 A0A0L0CF95 A0A1B0GD56 T1PKZ2 A0A1Q3FYV1 A0A182NQP4 A0A0P8ZWA1
PDB
1XNJ
E-value=3.26526e-43,
Score=434
Ontologies
GO
Topology
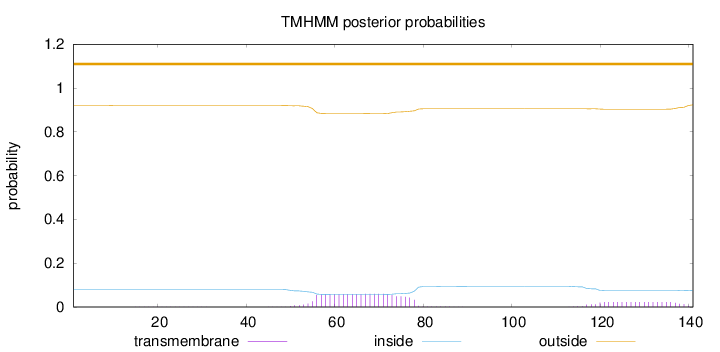
Length:
141
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.84532
Exp number, first 60 AAs:
0.36709
Total prob of N-in:
0.08006
outside
1 - 141
Population Genetic Test Statistics
Pi
268.207776
Theta
198.229957
Tajima's D
0.035059
CLR
0
CSRT
0.382130893455327
Interpretation
Uncertain
