Gene
KWMTBOMO12938 Validated by peptides from experiments
Annotation
PREDICTED:_27_kDa_glycoprotein-like_[Bombyx_mori]
Full name
27 kDa glycoprotein
+ More
27 kDa hemolymph protein
27 kDa hemolymph protein
Alternative Name
P27K
Location in the cell
Extracellular Reliability : 3.215
Sequence
CDS
ATGAATTTAAAAGTTTTGATTTTGTCTGCTATTATTGGAACTGCATATGCTCAGTCTGAGAAAGCTCTACTGGATAGTTTAAACTTGCCTCCTGAACTGCTGAACAATTTGAATGAGACTACTCTCCAAGAACATCAGAATAAAACCACAGAAGTGTTTAAAAATAAATGTGAACAAAATGGAGGACCTGAAGCTTTTAATAACGCACATAATGCATTCGATCAGTTAACATCATGCCTGCAAACCCTTGTGAATATGGAGATACTTCAGCAAGAGTTAGAGGAAGCTAAACCGAATGGAATGATCGATGAAGTCTTCAAAAAGTACTGCTTAAAGAAGACAAACTTCATGAATTGTTTCACAACTATGCTGGATGCTTCTAAGCCGTGCTTTACAGTTAAAGAGCGAGATCATCTCAACACAGTGTACAATGTGTCCCGAGAACTGGCTGACTTCGTTTGCTTCAAAGACGGTGATAGAATTGCATTGTTCATCTCTGAAGGAGGAGCGGAATGTTTTCAAGAGAAGCAGTCTGACTTGCAGAAGTGCTACAATCAGACAGCAGGGCAGAACGCTAACTTTGATTTGAACTCTTTATCTCCCGACAATTTACCAACTTTATCCTTTGATGAGAAGCAATGCAAGGAGATGTCCGTAACGCAAGTTTGCGTGGTGAAGGTGCTGGAGACGTGCCAGAAGCCGGCCGCCGCTAACATCGTCGACTCGCTCTTCAAGTTCCTGGTCAAGAAGACTCCCTGCAAGGCGTATCTGAAAGCTGAGGAGAATGCCCCCGGTTCTGCCGACGTGTTGTTCGCGAAAATGGCTACTGTCGCAATGGGACTTTTGGCCGTTGCTCTAGTCTAG
Protein
MNLKVLILSAIIGTAYAQSEKALLDSLNLPPELLNNLNETTLQEHQNKTTEVFKNKCEQNGGPEAFNNAHNAFDQLTSCLQTLVNMEILQQELEEAKPNGMIDEVFKKYCLKKTNFMNCFTTMLDASKPCFTVKERDHLNTVYNVSRELADFVCFKDGDRIALFISEGGAECFQEKQSDLQKCYNQTAGQNANFDLNSLSPDNLPTLSFDEKQCKEMSVTQVCVVKVLETCQKPAAANIVDSLFKFLVKKTPCKAYLKAEENAPGSADVLFAKMATVAMGLLAVALV
Summary
Subunit
Monomer.
Similarity
Belongs to the UPF0408 family.
Keywords
Complete proteome
Direct protein sequencing
Glycoprotein
Reference proteome
Secreted
Signal
Feature
chain 27 kDa glycoprotein
Uniprot
H9ISK6
S4PC68
A0A1E1WHQ2
A0A212F195
A0A194Q7Q0
A0A2J7PR17
+ More
E2C9D0 A0A0J7L587 A0A1B6D3A7 A0A0A9YLF7 A0A2A3EFD7 V9IIW7 A0A0K8SEY2 A0A026WU00 E2AMV6 R4G2X4 R4WPP9 A0A0C9QVB6 A0A0C9RF57 A0A0C9RBC0 A0A1J1J4R8 E9IFE0 A0A336L8A3 A0A336N5I4 K7J4H8 D6WZA1 A0A087ZSL8 A0A1B6LB09 A0A1L8DP37 I4DRA5 A0A224XTU8 A0A1B6FPP0 A0A1B6MAC1 A0A232EN48 A0A0N0PBD7 V5GLV5 A0A151WRK4 A0A1B6HV20 A0A1B6H6W4 A0A151IJS8 B4NCM3 A0A1Q3FR51 I4DM97 A0A2H1VXC7 B0VZS6 Q8T113 Q95SC0 Q9VXH7 B4IF58 A0A1W4WB54 B4R5S5 A0A158NKK7 B4M287 B4PXR5 I4DIP5 A0A023EQE4 A0A182H6H9 A0A182T1H9 A0A2S2P679 A0A2A4J1E9 A0A182MVI0 A0A023EN33 A0A023EN21 A0A023EP79 A0A3S2TQN3 A0A182GAJ9 A0A182GAK0 A0A1Q3FP95 A0A023EMF7 A0A1S3DEU1 B3NX57 A0A151JB59 U5ESG4 A0A182VE44 A0A212F1C5 A0A182TGD7 Q7PTC7 A0A182KRX8 Q16YP3 A0A2M4ARB3 B0VZS5 A0A1B6GKM1 A0A2H8U2X3 A0A2C9GRV1 A0A2H4N0S2 A0A3B0JWY4 A0A2M4APP8 A0A1W4W214 A0A2M4AQK1 A0A182MWE9 J9JPJ9 S4NPG4 P83632 Q29GE6 A0A182XZG0
E2C9D0 A0A0J7L587 A0A1B6D3A7 A0A0A9YLF7 A0A2A3EFD7 V9IIW7 A0A0K8SEY2 A0A026WU00 E2AMV6 R4G2X4 R4WPP9 A0A0C9QVB6 A0A0C9RF57 A0A0C9RBC0 A0A1J1J4R8 E9IFE0 A0A336L8A3 A0A336N5I4 K7J4H8 D6WZA1 A0A087ZSL8 A0A1B6LB09 A0A1L8DP37 I4DRA5 A0A224XTU8 A0A1B6FPP0 A0A1B6MAC1 A0A232EN48 A0A0N0PBD7 V5GLV5 A0A151WRK4 A0A1B6HV20 A0A1B6H6W4 A0A151IJS8 B4NCM3 A0A1Q3FR51 I4DM97 A0A2H1VXC7 B0VZS6 Q8T113 Q95SC0 Q9VXH7 B4IF58 A0A1W4WB54 B4R5S5 A0A158NKK7 B4M287 B4PXR5 I4DIP5 A0A023EQE4 A0A182H6H9 A0A182T1H9 A0A2S2P679 A0A2A4J1E9 A0A182MVI0 A0A023EN33 A0A023EN21 A0A023EP79 A0A3S2TQN3 A0A182GAJ9 A0A182GAK0 A0A1Q3FP95 A0A023EMF7 A0A1S3DEU1 B3NX57 A0A151JB59 U5ESG4 A0A182VE44 A0A212F1C5 A0A182TGD7 Q7PTC7 A0A182KRX8 Q16YP3 A0A2M4ARB3 B0VZS5 A0A1B6GKM1 A0A2H8U2X3 A0A2C9GRV1 A0A2H4N0S2 A0A3B0JWY4 A0A2M4APP8 A0A1W4W214 A0A2M4AQK1 A0A182MWE9 J9JPJ9 S4NPG4 P83632 Q29GE6 A0A182XZG0
Pubmed
19121390
23622113
22118469
26354079
20798317
25401762
+ More
26823975 24508170 30249741 23691247 21282665 20075255 18362917 19820115 22651552 28648823 17994087 11280994 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 17550304 24945155 26483478 12364791 14747013 17210077 20966253 17510324 15632085 25244985
26823975 24508170 30249741 23691247 21282665 20075255 18362917 19820115 22651552 28648823 17994087 11280994 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21347285 17550304 24945155 26483478 12364791 14747013 17210077 20966253 17510324 15632085 25244985
EMBL
BABH01013486
BABH01013487
GAIX01004301
JAA88259.1
GDQN01009790
GDQN01004530
+ More
JAT81264.1 JAT86524.1 AGBW02010923 OWR47512.1 KQ459324 KPJ01562.1 NEVH01022634 PNF18756.1 GL453806 EFN75475.1 LBMM01000718 KMQ97649.1 GEDC01017127 JAS20171.1 GBHO01013234 GDHC01005733 JAG30370.1 JAQ12896.1 KZ288266 PBC30194.1 JR047553 AEY60441.1 GBRD01014167 JAG51659.1 KK107105 QOIP01000011 EZA59545.1 RLU16634.1 GL440936 EFN65230.1 GAHY01002206 JAA75304.1 AK417641 BAN20856.1 GBYB01004617 JAG74384.1 GBYB01005621 JAG75388.1 GBYB01005620 JAG75387.1 CVRI01000070 CRL07400.1 GL762838 EFZ20728.1 UFQS01002272 UFQT01002272 SSX13634.1 SSX33060.1 UFQT01002606 SSX33758.1 KQ971372 EFA09742.1 GEBQ01019085 JAT20892.1 GFDF01005947 JAV08137.1 AK405040 BAM20445.1 GFTR01004484 JAW11942.1 GECZ01017607 GECZ01004595 JAS52162.1 JAS65174.1 GEBQ01007094 GEBQ01005295 GEBQ01000335 JAT32883.1 JAT34682.1 JAT39642.1 NNAY01003237 OXU19756.1 KQ460968 KPJ10218.1 GALX01003417 JAB65049.1 KQ982805 KYQ50496.1 GECU01029172 GECU01014812 JAS78534.1 JAS92894.1 GECU01037335 GECU01033259 JAS70371.1 JAS74447.1 KQ977294 KYN03883.1 CH964239 EDW82582.2 GFDL01004945 JAV30100.1 AK402415 BAM19037.1 ODYU01005014 SOQ45485.1 DS231815 EDS35168.1 AB062103 BAB87849.1 AY060868 BT072832 AAL28416.1 ACN62435.2 AE014298 BT132979 AAF48590.1 AEW68191.1 AGB95447.1 CH480832 EDW46112.1 CM000366 EDX18075.1 ADTU01018871 ADTU01018872 CH940651 EDW65791.1 CM000162 EDX01901.1 AK401163 BAM17785.1 KPJ01561.1 GAPW01002809 JAC10789.1 JXUM01114166 KQ565760 KXJ70685.1 GGMR01012243 MBY24862.1 NWSH01003653 PCG65987.1 AXCM01000560 GAPW01002811 JAC10787.1 GAPW01002810 JAC10788.1 JXUM01114165 GAPW01002812 JAC10786.1 KXJ70684.1 RSAL01000021 RVE52498.1 JXUM01008680 KQ560266 KXJ83463.1 JXUM01008681 KXJ83464.1 GFDL01005701 JAV29344.1 GAPW01003578 JAC10020.1 CH954180 EDV46886.1 KQ979182 KYN22310.1 GANO01003257 JAB56614.1 OWR47513.1 AAAB01008807 EAA04107.3 CH477513 EAT39730.1 GGFK01010015 MBW43336.1 EDS35167.1 GECZ01006773 JAS62996.1 GFXV01008123 MBW19928.1 APCN01000220 KY653090 ATU47284.1 OUUW01000011 SPP86597.1 GGFK01009450 MBW42771.1 GGFK01009745 MBW43066.1 ABLF02041139 GAIX01011874 JAA80686.1 AJ575661 CAE02611.1 CH379064 EAL32163.1
JAT81264.1 JAT86524.1 AGBW02010923 OWR47512.1 KQ459324 KPJ01562.1 NEVH01022634 PNF18756.1 GL453806 EFN75475.1 LBMM01000718 KMQ97649.1 GEDC01017127 JAS20171.1 GBHO01013234 GDHC01005733 JAG30370.1 JAQ12896.1 KZ288266 PBC30194.1 JR047553 AEY60441.1 GBRD01014167 JAG51659.1 KK107105 QOIP01000011 EZA59545.1 RLU16634.1 GL440936 EFN65230.1 GAHY01002206 JAA75304.1 AK417641 BAN20856.1 GBYB01004617 JAG74384.1 GBYB01005621 JAG75388.1 GBYB01005620 JAG75387.1 CVRI01000070 CRL07400.1 GL762838 EFZ20728.1 UFQS01002272 UFQT01002272 SSX13634.1 SSX33060.1 UFQT01002606 SSX33758.1 KQ971372 EFA09742.1 GEBQ01019085 JAT20892.1 GFDF01005947 JAV08137.1 AK405040 BAM20445.1 GFTR01004484 JAW11942.1 GECZ01017607 GECZ01004595 JAS52162.1 JAS65174.1 GEBQ01007094 GEBQ01005295 GEBQ01000335 JAT32883.1 JAT34682.1 JAT39642.1 NNAY01003237 OXU19756.1 KQ460968 KPJ10218.1 GALX01003417 JAB65049.1 KQ982805 KYQ50496.1 GECU01029172 GECU01014812 JAS78534.1 JAS92894.1 GECU01037335 GECU01033259 JAS70371.1 JAS74447.1 KQ977294 KYN03883.1 CH964239 EDW82582.2 GFDL01004945 JAV30100.1 AK402415 BAM19037.1 ODYU01005014 SOQ45485.1 DS231815 EDS35168.1 AB062103 BAB87849.1 AY060868 BT072832 AAL28416.1 ACN62435.2 AE014298 BT132979 AAF48590.1 AEW68191.1 AGB95447.1 CH480832 EDW46112.1 CM000366 EDX18075.1 ADTU01018871 ADTU01018872 CH940651 EDW65791.1 CM000162 EDX01901.1 AK401163 BAM17785.1 KPJ01561.1 GAPW01002809 JAC10789.1 JXUM01114166 KQ565760 KXJ70685.1 GGMR01012243 MBY24862.1 NWSH01003653 PCG65987.1 AXCM01000560 GAPW01002811 JAC10787.1 GAPW01002810 JAC10788.1 JXUM01114165 GAPW01002812 JAC10786.1 KXJ70684.1 RSAL01000021 RVE52498.1 JXUM01008680 KQ560266 KXJ83463.1 JXUM01008681 KXJ83464.1 GFDL01005701 JAV29344.1 GAPW01003578 JAC10020.1 CH954180 EDV46886.1 KQ979182 KYN22310.1 GANO01003257 JAB56614.1 OWR47513.1 AAAB01008807 EAA04107.3 CH477513 EAT39730.1 GGFK01010015 MBW43336.1 EDS35167.1 GECZ01006773 JAS62996.1 GFXV01008123 MBW19928.1 APCN01000220 KY653090 ATU47284.1 OUUW01000011 SPP86597.1 GGFK01009450 MBW42771.1 GGFK01009745 MBW43066.1 ABLF02041139 GAIX01011874 JAA80686.1 AJ575661 CAE02611.1 CH379064 EAL32163.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000235965
UP000008237
UP000036403
+ More
UP000242457 UP000053097 UP000279307 UP000000311 UP000183832 UP000002358 UP000007266 UP000005203 UP000215335 UP000053240 UP000075809 UP000078542 UP000007798 UP000002320 UP000000803 UP000001292 UP000192223 UP000000304 UP000005205 UP000008792 UP000002282 UP000069940 UP000249989 UP000075901 UP000218220 UP000075883 UP000283053 UP000079169 UP000008711 UP000078492 UP000075903 UP000075902 UP000007062 UP000075882 UP000008820 UP000075840 UP000268350 UP000192221 UP000007819 UP000001819 UP000076408
UP000242457 UP000053097 UP000279307 UP000000311 UP000183832 UP000002358 UP000007266 UP000005203 UP000215335 UP000053240 UP000075809 UP000078542 UP000007798 UP000002320 UP000000803 UP000001292 UP000192223 UP000000304 UP000005205 UP000008792 UP000002282 UP000069940 UP000249989 UP000075901 UP000218220 UP000075883 UP000283053 UP000079169 UP000008711 UP000078492 UP000075903 UP000075902 UP000007062 UP000075882 UP000008820 UP000075840 UP000268350 UP000192221 UP000007819 UP000001819 UP000076408
Interpro
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
H9ISK6
S4PC68
A0A1E1WHQ2
A0A212F195
A0A194Q7Q0
A0A2J7PR17
+ More
E2C9D0 A0A0J7L587 A0A1B6D3A7 A0A0A9YLF7 A0A2A3EFD7 V9IIW7 A0A0K8SEY2 A0A026WU00 E2AMV6 R4G2X4 R4WPP9 A0A0C9QVB6 A0A0C9RF57 A0A0C9RBC0 A0A1J1J4R8 E9IFE0 A0A336L8A3 A0A336N5I4 K7J4H8 D6WZA1 A0A087ZSL8 A0A1B6LB09 A0A1L8DP37 I4DRA5 A0A224XTU8 A0A1B6FPP0 A0A1B6MAC1 A0A232EN48 A0A0N0PBD7 V5GLV5 A0A151WRK4 A0A1B6HV20 A0A1B6H6W4 A0A151IJS8 B4NCM3 A0A1Q3FR51 I4DM97 A0A2H1VXC7 B0VZS6 Q8T113 Q95SC0 Q9VXH7 B4IF58 A0A1W4WB54 B4R5S5 A0A158NKK7 B4M287 B4PXR5 I4DIP5 A0A023EQE4 A0A182H6H9 A0A182T1H9 A0A2S2P679 A0A2A4J1E9 A0A182MVI0 A0A023EN33 A0A023EN21 A0A023EP79 A0A3S2TQN3 A0A182GAJ9 A0A182GAK0 A0A1Q3FP95 A0A023EMF7 A0A1S3DEU1 B3NX57 A0A151JB59 U5ESG4 A0A182VE44 A0A212F1C5 A0A182TGD7 Q7PTC7 A0A182KRX8 Q16YP3 A0A2M4ARB3 B0VZS5 A0A1B6GKM1 A0A2H8U2X3 A0A2C9GRV1 A0A2H4N0S2 A0A3B0JWY4 A0A2M4APP8 A0A1W4W214 A0A2M4AQK1 A0A182MWE9 J9JPJ9 S4NPG4 P83632 Q29GE6 A0A182XZG0
E2C9D0 A0A0J7L587 A0A1B6D3A7 A0A0A9YLF7 A0A2A3EFD7 V9IIW7 A0A0K8SEY2 A0A026WU00 E2AMV6 R4G2X4 R4WPP9 A0A0C9QVB6 A0A0C9RF57 A0A0C9RBC0 A0A1J1J4R8 E9IFE0 A0A336L8A3 A0A336N5I4 K7J4H8 D6WZA1 A0A087ZSL8 A0A1B6LB09 A0A1L8DP37 I4DRA5 A0A224XTU8 A0A1B6FPP0 A0A1B6MAC1 A0A232EN48 A0A0N0PBD7 V5GLV5 A0A151WRK4 A0A1B6HV20 A0A1B6H6W4 A0A151IJS8 B4NCM3 A0A1Q3FR51 I4DM97 A0A2H1VXC7 B0VZS6 Q8T113 Q95SC0 Q9VXH7 B4IF58 A0A1W4WB54 B4R5S5 A0A158NKK7 B4M287 B4PXR5 I4DIP5 A0A023EQE4 A0A182H6H9 A0A182T1H9 A0A2S2P679 A0A2A4J1E9 A0A182MVI0 A0A023EN33 A0A023EN21 A0A023EP79 A0A3S2TQN3 A0A182GAJ9 A0A182GAK0 A0A1Q3FP95 A0A023EMF7 A0A1S3DEU1 B3NX57 A0A151JB59 U5ESG4 A0A182VE44 A0A212F1C5 A0A182TGD7 Q7PTC7 A0A182KRX8 Q16YP3 A0A2M4ARB3 B0VZS5 A0A1B6GKM1 A0A2H8U2X3 A0A2C9GRV1 A0A2H4N0S2 A0A3B0JWY4 A0A2M4APP8 A0A1W4W214 A0A2M4AQK1 A0A182MWE9 J9JPJ9 S4NPG4 P83632 Q29GE6 A0A182XZG0
Ontologies
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 17,
Likelihood: 0.985393
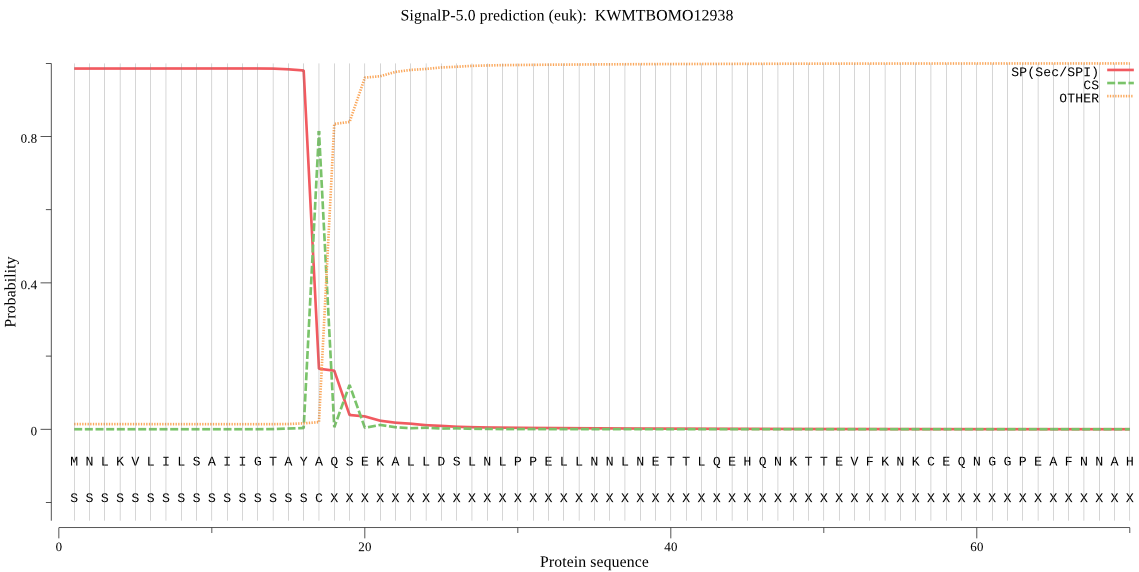
Length:
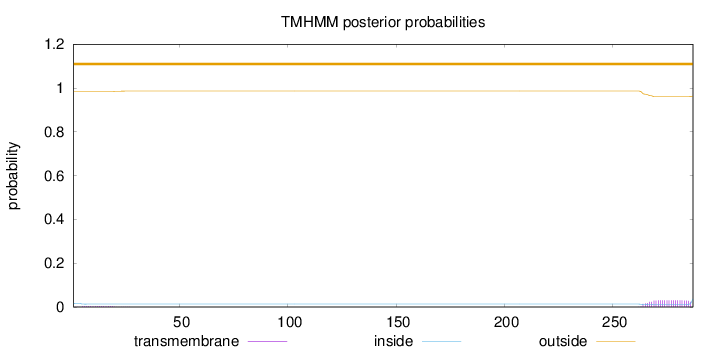
287
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.69399
Exp number, first 60 AAs:
0.06665
Total prob of N-in:
0.01703
outside
1 - 287
Population Genetic Test Statistics
Pi
148.936081
Theta
172.021013
Tajima's D
0.199283
CLR
208.425966
CSRT
0.421078946052697
Interpretation
Uncertain
