Gene
KWMTBOMO12931
Pre Gene Modal
BGIBMGA000467
Annotation
PREDICTED:_protocadherin-like_wing_polarity_protein_stan_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.833 PlasmaMembrane Reliability : 2.054
Sequence
CDS
ATGAACACGTTTGTCGGGGTCCGCGTGGCCGCCGACCTGAGCACCGCGTGCGCCAACACGCCGCGCCTCTACGGAGCCGACGTCGTGGTCGCCGAGGGAATACTGCTCGAGCTCATGGACTACGAGCTGAGACAAGCCGGCTTGAACCTTACGCACAGTCAAGATAAGGACTATGTTCGGAATTTACTGAAATCAACGAACACAATACTAAACGTCGAATACGAGAGAGAATGGAAACGCATCCGCGAGCTGACGGGCCACGGCGCGGAACTGCTCGTGCAGAAGTTCGACGAGTACATAGCGGTGCTCGCCGAGAGCCAACACGACACCTACACCAGCCCCTTCGAGATCGTCACTAAGGACGTGGTGATCAGCGTGGACACAGTGACTTCGGAGTCCATTTACGGCTTCGAACCGACACAGTTGAACCGCCTCAACGTCGACTCCTACACGACCGAGAGAGTGGTCGCTCTGCCTGACACGTCGGCGTTCATACATTCGCCCATACAGGCAACCGGTTACTATGGCAACGGGAAAACTAACAAAAACAAGAAACCTTCCAGCCCCACCGTGTCCTTCCCGAAGTACAATAATTACGTGAGGAGCAAGAATAAGTTCGACAAATACACTAGAGTATTGCTTCCTTTGGATCTGTTGGGAATTTCAAATAATAATCTTCAAGACATAACGGACACGAACTACGAAACGCGCGCCGTGTTCGGCTACGTCCAGTACTCGAGGAATTCGTCTCAACTGATGCCCCTGCGCATGGACGACTCCGTCAGTCTTCGATGGGGCGTCAACATCACAATAGGATCGCCGATATTGCAAATAGCAGTCTTCGTACCGGAATCCATCTGGACCAGAGAGACCGGTGAAGGAAGCGAGGATAATTCCATCGAGCACAATGCTGAAGAAACCGAAGGCATCCTTTACGCAAACAGAGGCACAAAGCATTACCAGGCTAACCGCGTTAGCAGCCAGCGCCAGAACCGGCCACACACAAAAGAAGATCCTCCCGCGGAACACGGCCCCGTGCTCATACAACCCGCGTTCAAAAAACACGAAAGAAACGACCAGATACAGGAAAACGAGTCGTTGTTTGTCGCCGATAGAGACGACGGACCCAAAGTGATGGCGTTGGTCATGGACGGAGAGACCAGAAGCAACGCGACCGGTGTCGGCAAGAAGATGTACAGAAGTCTCAGCGGTATAAGACTGAAGAAGCCGATCAGACTACAGATTTGGCTCGATATAAACAGAGAAACGTTCGGTTCGAGAACTAACCCGCAATGTGTGCACTGGTCGACATTGAGAGGGTCCGGTGAATGGTCTCGCGTCGGTTGTCACACAGAAATAGACTACGATTGGTCGCCGTACTCTGAAGAGCCGCTACTAATCAACTGCACGTGCAATCATTTGTCTACGTTTGCCGTTCTCGTCGACGAAGTGGACGTTGAGTTCATTCCTGAGCCGTCCCTGCTCGAGTCAGTGACGTCATACACTGCATTCAGTATATCGCTCCCGCTCTTGCTCACGTCATGGGCCGCGCTCTGTCTCATTCGCGGGGGGGCTGCCACAGTATCGAACTCGATACACAAACATTTGATATTCAGCGTGTTTATGGCCGAGTTATTGTATTTGATCGCACTGAAGGCGCGAACGTCTTTAGTCCAGAATGAGTTCGCGTGCAAGGTGGTGGCGATGTCGCTGCACTACTGGTGGGTGTGCGGCGCGGGCTGGGCGGCGTGCGACGCGTGGCAGCTGCGCCGGCTGCTGCGCGAGCTGCGCGACGTCAACCACGCGCCGCTGTGCGCGCCGCTCGCCGCCGCCTACGCCGCGCCCGCACTCGTGCTCGCGCTCGCCGCCTCGCACCGCCCGCACCAGTACGGGAACGCGCTCTTTTGCTGGGTGAGCTGCCACGAGCCCGTGGTGTGGTGGGTGGTGATCCCGGCGGGAGTGTACGCGGCGGCCGCCTTCGTGTTCCTCGCCGGAGCCACCAGGGCGGCCTTCACGGTCAGGGACCACGTCACGGGATTCGGCAACTTGAGGATGCTGCTAGCGGTCAGCATCGTGTGTCTCCCTCTGCTGTGCGTGGCCTGGGCGAGCGCGTTGCTGCTGGGTAGCGAGGCGGGGGGCCGTCGCCACGCGCTCAGCGCCGTGCTGTCCGGCGCCGTATTGCTCCACGCCATCGCCGCTTCCCTCGGATACATCGCCTTCAACGTCCGCGTACGGGACAACTTGAAGAGGACGGTATTAAGGTGCATGGGGAAGAAAGTGCCTTTAGCCGATACGTCTGTCATAATCAGCAGCAGTGCCCATAACCTCTCACAAGTGAACAGATCATCATTGGCTTACCACAGCAGCACGGAGCCAGGTCGGCAGATGCGGAACATTGGCATTTCAGCGTCTTCCACGACATCTAGATCCACGACGAAAACTAGTTCTAGTCCATACAGCCGCAGCGATGGACAGCTACGTCACACCAGCACCTCGACGTCTAACTACAACACGAGCGCGAGCGACATGCCCGCCTACCTCCGGGGATTCGACTCCTCGTTACACACCAGAAAAGACGATGAGCGTCATCGGCGCAAGTCGGAGAGGTCGGAGAGGGGTACGGAGCGGGGGGAGCGCGGGGAGGGCACGGAGGGGAGCGACAGTGAGAGCGAGGGCAGCGCGCGGAGCCTCGAACTCGCCTCCAGCCACTCGTCAGACGACGACGACACCTCCACCAGGAGATCCAGCCACCCCCCCTCTGCCAACAGAGAGCGCATCGCCAGCGGAAGTGCCAACAGCTATTTACCAAATATAACGGAAGGCGCTCCTTTGGCTATGTCCCCAAGATGGCCGTCTCACGTGAGAGAAGACCAGAGCCGGCCGGAAATGGGTAGATGGTCGCAGGAGACCGGCTCCGACAACGAGGGCGCGAACATGGGCATCGCGACGCCATCGCCCAACCCGCTCCCCAACCCGGACCTCACGTCCGACGTGTACCTCAACCGGCACCGCCTCAAGCCGTCGATATTGGAGAACGTACACGACACCTATGTCGCTAGTGTCGACGCTTCGAGACTGCCTCTTGATAACAGGTACGGAGTGATGGAAGACGAATTAATGTACGGTGTTTTACCTATCGATCGAGAAAAACAAATGCCATACGATCCGAACTACCCGATATCCCCCACGATGTACCGGCTACCCGGGAACCCCTACGGCTCCAAAACCTACCTATCGTCGAGAACTTCGGATTACGGGTACAGTCCAACGAAATACATGAACGATACGAACGTATACGGGTCGAGGGATTTCCGTGACTCCGGGGTCTCGACGAGGGACCACGACGGCTACCCGCCCAGGGACTTCAGGGACTCTGGTATAGCTACGATGCTGAAGAACGACTATCAGAGGAAAATGGATGGGCACTACGGTGAATACAACGACCGGATGTCCGAGGGATCCGAAAAACATCACGGGCACGACAAATACTTGTTCCCGTACACGGCCGAGGAAGATCACGCAACTTTGCGTGGCTTGAGTCAGTCTCCTCGCACGCGTAGTGAGATCCAACATGCAGGGGAAGCACCACAGCAGATGGGGGCGCCACTGGCAAGCATGGGGGAAACAAGCGAGAAATCTACTACTGAAGACGACGCCGAAACAAGAGTATAA
Protein
MNTFVGVRVAADLSTACANTPRLYGADVVVAEGILLELMDYELRQAGLNLTHSQDKDYVRNLLKSTNTILNVEYEREWKRIRELTGHGAELLVQKFDEYIAVLAESQHDTYTSPFEIVTKDVVISVDTVTSESIYGFEPTQLNRLNVDSYTTERVVALPDTSAFIHSPIQATGYYGNGKTNKNKKPSSPTVSFPKYNNYVRSKNKFDKYTRVLLPLDLLGISNNNLQDITDTNYETRAVFGYVQYSRNSSQLMPLRMDDSVSLRWGVNITIGSPILQIAVFVPESIWTRETGEGSEDNSIEHNAEETEGILYANRGTKHYQANRVSSQRQNRPHTKEDPPAEHGPVLIQPAFKKHERNDQIQENESLFVADRDDGPKVMALVMDGETRSNATGVGKKMYRSLSGIRLKKPIRLQIWLDINRETFGSRTNPQCVHWSTLRGSGEWSRVGCHTEIDYDWSPYSEEPLLINCTCNHLSTFAVLVDEVDVEFIPEPSLLESVTSYTAFSISLPLLLTSWAALCLIRGGAATVSNSIHKHLIFSVFMAELLYLIALKARTSLVQNEFACKVVAMSLHYWWVCGAGWAACDAWQLRRLLRELRDVNHAPLCAPLAAAYAAPALVLALAASHRPHQYGNALFCWVSCHEPVVWWVVIPAGVYAAAAFVFLAGATRAAFTVRDHVTGFGNLRMLLAVSIVCLPLLCVAWASALLLGSEAGGRRHALSAVLSGAVLLHAIAASLGYIAFNVRVRDNLKRTVLRCMGKKVPLADTSVIISSSAHNLSQVNRSSLAYHSSTEPGRQMRNIGISASSTTSRSTTKTSSSPYSRSDGQLRHTSTSTSNYNTSASDMPAYLRGFDSSLHTRKDDERHRRKSERSERGTERGERGEGTEGSDSESEGSARSLELASSHSSDDDDTSTRRSSHPPSANRERIASGSANSYLPNITEGAPLAMSPRWPSHVREDQSRPEMGRWSQETGSDNEGANMGIATPSPNPLPNPDLTSDVYLNRHRLKPSILENVHDTYVASVDASRLPLDNRYGVMEDELMYGVLPIDREKQMPYDPNYPISPTMYRLPGNPYGSKTYLSSRTSDYGYSPTKYMNDTNVYGSRDFRDSGVSTRDHDGYPPRDFRDSGIATMLKNDYQRKMDGHYGEYNDRMSEGSEKHHGHDKYLFPYTAEEDHATLRGLSQSPRTRSEIQHAGEAPQQMGAPLASMGETSEKSTTEDDAETRV
Summary
Similarity
Belongs to the G-protein coupled receptor 2 family.
Uniprot
A0A2H1W4W7
A0A3S2M6G4
A0A194Q7S2
A0A0N0PBD5
A0A2A4J4K8
A0A2A4J5X8
+ More
A0A0K8UWC5 A0A0K8TWT7 A0A1B0G6P0 A0A1A9VY21 A0A1A9ZX75 A0A1B0C2G8 A0A1A9YB23 A0A1B0BHL9 A0A1A9WYK3 B4P848 A0A1J1HPP8 B4HMT2 Q17PS6 A0A1W4XB87 A0A1S4EVA6 A0A0M4EEV1 A0A336MHS2 A0A336MVD4 A0A182NV34 D6X315 A0A139WBL5 A0A182JQ17 A0A1B0CFP2 A0A182RQF7 A0A182WMK2 A0A182VDC9 Q7PPU8 A0A182WS38 A0A182LHX3 A0A182P0K3 A0A182QFS4 A0A182MR91 A0A182U1V4 A0A182HXT4 W5JQ54 A0A182YBV7 U4UCN9 A0A1Y1KMS4 A0A182FCS1 A0A182G8Q4 A0A0T6AXN9 B0XD21 A0A1S4KAM1 A0A182SZM8 A0A1I8MRY5 A0A1I8NR16 A0A1I8NR14 A0A1I8NR21 A0A084VUD4
A0A0K8UWC5 A0A0K8TWT7 A0A1B0G6P0 A0A1A9VY21 A0A1A9ZX75 A0A1B0C2G8 A0A1A9YB23 A0A1B0BHL9 A0A1A9WYK3 B4P848 A0A1J1HPP8 B4HMT2 Q17PS6 A0A1W4XB87 A0A1S4EVA6 A0A0M4EEV1 A0A336MHS2 A0A336MVD4 A0A182NV34 D6X315 A0A139WBL5 A0A182JQ17 A0A1B0CFP2 A0A182RQF7 A0A182WMK2 A0A182VDC9 Q7PPU8 A0A182WS38 A0A182LHX3 A0A182P0K3 A0A182QFS4 A0A182MR91 A0A182U1V4 A0A182HXT4 W5JQ54 A0A182YBV7 U4UCN9 A0A1Y1KMS4 A0A182FCS1 A0A182G8Q4 A0A0T6AXN9 B0XD21 A0A1S4KAM1 A0A182SZM8 A0A1I8MRY5 A0A1I8NR16 A0A1I8NR14 A0A1I8NR21 A0A084VUD4
Pubmed
EMBL
ODYU01006320
SOQ48063.1
RSAL01000021
RVE52505.1
KQ459324
KPJ01568.1
+ More
KQ460968 KPJ10225.1 NWSH01003030 PCG67077.1 PCG67078.1 GDHF01021433 JAI30881.1 GDHF01033593 JAI18721.1 CCAG010015976 JXJN01024530 JXJN01014429 CM000158 EDW90095.2 CVRI01000015 CRK90013.1 CH480816 EDW47294.1 CH477189 EAT48765.2 CP012524 ALC40622.1 UFQT01001209 SSX29510.1 UFQT01001301 SSX29908.1 KQ971372 EFA10758.2 KYB25255.1 AJWK01010222 AAAB01008905 EAA09721.2 AXCN02001921 AXCM01004264 APCN01005346 ADMH02000839 ETN64889.1 KB632203 ERL90083.1 GEZM01078449 JAV62703.1 JXUM01152499 KQ571471 KXJ68168.1 LJIG01022594 KRT79748.1 DS232735 EDS45256.1 ATLV01016720 KE525103 KFB41578.1
KQ460968 KPJ10225.1 NWSH01003030 PCG67077.1 PCG67078.1 GDHF01021433 JAI30881.1 GDHF01033593 JAI18721.1 CCAG010015976 JXJN01024530 JXJN01014429 CM000158 EDW90095.2 CVRI01000015 CRK90013.1 CH480816 EDW47294.1 CH477189 EAT48765.2 CP012524 ALC40622.1 UFQT01001209 SSX29510.1 UFQT01001301 SSX29908.1 KQ971372 EFA10758.2 KYB25255.1 AJWK01010222 AAAB01008905 EAA09721.2 AXCN02001921 AXCM01004264 APCN01005346 ADMH02000839 ETN64889.1 KB632203 ERL90083.1 GEZM01078449 JAV62703.1 JXUM01152499 KQ571471 KXJ68168.1 LJIG01022594 KRT79748.1 DS232735 EDS45256.1 ATLV01016720 KE525103 KFB41578.1
Proteomes
UP000283053
UP000053268
UP000053240
UP000218220
UP000092444
UP000078200
+ More
UP000092445 UP000092460 UP000092443 UP000091820 UP000002282 UP000183832 UP000001292 UP000008820 UP000192223 UP000092553 UP000075884 UP000007266 UP000075881 UP000092461 UP000075900 UP000075920 UP000075903 UP000007062 UP000076407 UP000075882 UP000075885 UP000075886 UP000075883 UP000075902 UP000075840 UP000000673 UP000076408 UP000030742 UP000069272 UP000069940 UP000249989 UP000002320 UP000075901 UP000095301 UP000095300 UP000030765
UP000092445 UP000092460 UP000092443 UP000091820 UP000002282 UP000183832 UP000001292 UP000008820 UP000192223 UP000092553 UP000075884 UP000007266 UP000075881 UP000092461 UP000075900 UP000075920 UP000075903 UP000007062 UP000076407 UP000075882 UP000075885 UP000075886 UP000075883 UP000075902 UP000075840 UP000000673 UP000076408 UP000030742 UP000069272 UP000069940 UP000249989 UP000002320 UP000075901 UP000095301 UP000095300 UP000030765
Pfam
Interpro
IPR013032
EGF-like_CS
+ More
IPR036445 GPCR_2_extracell_dom_sf
IPR000742 EGF-like_dom
IPR001791 Laminin_G
IPR017981 GPCR_2-like
IPR000832 GPCR_2_secretin-like
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR032471 GAIN_dom_N
IPR001881 EGF-like_Ca-bd_dom
IPR013320 ConA-like_dom_sf
IPR001879 GPCR_2_extracellular_dom
IPR002049 Laminin_EGF
IPR000203 GPS
IPR002126 Cadherin-like_dom
IPR020894 Cadherin_CS
IPR015919 Cadherin-like_sf
IPR009030 Growth_fac_rcpt_cys_sf
IPR036445 GPCR_2_extracell_dom_sf
IPR000742 EGF-like_dom
IPR001791 Laminin_G
IPR017981 GPCR_2-like
IPR000832 GPCR_2_secretin-like
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR032471 GAIN_dom_N
IPR001881 EGF-like_Ca-bd_dom
IPR013320 ConA-like_dom_sf
IPR001879 GPCR_2_extracellular_dom
IPR002049 Laminin_EGF
IPR000203 GPS
IPR002126 Cadherin-like_dom
IPR020894 Cadherin_CS
IPR015919 Cadherin-like_sf
IPR009030 Growth_fac_rcpt_cys_sf
Gene 3D
ProteinModelPortal
A0A2H1W4W7
A0A3S2M6G4
A0A194Q7S2
A0A0N0PBD5
A0A2A4J4K8
A0A2A4J5X8
+ More
A0A0K8UWC5 A0A0K8TWT7 A0A1B0G6P0 A0A1A9VY21 A0A1A9ZX75 A0A1B0C2G8 A0A1A9YB23 A0A1B0BHL9 A0A1A9WYK3 B4P848 A0A1J1HPP8 B4HMT2 Q17PS6 A0A1W4XB87 A0A1S4EVA6 A0A0M4EEV1 A0A336MHS2 A0A336MVD4 A0A182NV34 D6X315 A0A139WBL5 A0A182JQ17 A0A1B0CFP2 A0A182RQF7 A0A182WMK2 A0A182VDC9 Q7PPU8 A0A182WS38 A0A182LHX3 A0A182P0K3 A0A182QFS4 A0A182MR91 A0A182U1V4 A0A182HXT4 W5JQ54 A0A182YBV7 U4UCN9 A0A1Y1KMS4 A0A182FCS1 A0A182G8Q4 A0A0T6AXN9 B0XD21 A0A1S4KAM1 A0A182SZM8 A0A1I8MRY5 A0A1I8NR16 A0A1I8NR14 A0A1I8NR21 A0A084VUD4
A0A0K8UWC5 A0A0K8TWT7 A0A1B0G6P0 A0A1A9VY21 A0A1A9ZX75 A0A1B0C2G8 A0A1A9YB23 A0A1B0BHL9 A0A1A9WYK3 B4P848 A0A1J1HPP8 B4HMT2 Q17PS6 A0A1W4XB87 A0A1S4EVA6 A0A0M4EEV1 A0A336MHS2 A0A336MVD4 A0A182NV34 D6X315 A0A139WBL5 A0A182JQ17 A0A1B0CFP2 A0A182RQF7 A0A182WMK2 A0A182VDC9 Q7PPU8 A0A182WS38 A0A182LHX3 A0A182P0K3 A0A182QFS4 A0A182MR91 A0A182U1V4 A0A182HXT4 W5JQ54 A0A182YBV7 U4UCN9 A0A1Y1KMS4 A0A182FCS1 A0A182G8Q4 A0A0T6AXN9 B0XD21 A0A1S4KAM1 A0A182SZM8 A0A1I8MRY5 A0A1I8NR16 A0A1I8NR14 A0A1I8NR21 A0A084VUD4
Ontologies
GO
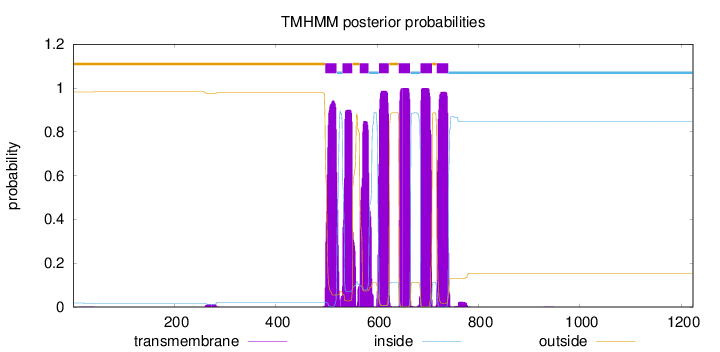
Topology
Length:
1223
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
146.39034
Exp number, first 60 AAs:
0.01134
Total prob of N-in:
0.01798
outside
1 - 497
TMhelix
498 - 520
inside
521 - 531
TMhelix
532 - 551
outside
552 - 565
TMhelix
566 - 583
inside
584 - 603
TMhelix
604 - 623
outside
624 - 642
TMhelix
643 - 665
inside
666 - 685
TMhelix
686 - 708
outside
709 - 717
TMhelix
718 - 740
inside
741 - 1223
Population Genetic Test Statistics
Pi
30.814654
Theta
198.298525
Tajima's D
2.037024
CLR
0.580064
CSRT
0.889955502224889
Interpretation
Uncertain
