Pre Gene Modal
BGIBMGA000472
Annotation
PREDICTED:_ATP-binding_cassette_sub-family_G_member_5_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.53
Sequence
CDS
ATGATCGGTGGCGATTATACTTTGGAGCTGTGCAACGTTTTCCATTCGGGACAGGTGGAACCAGGCAGCTTCTTCCAACGTCTCACTGGAGGCGTGAAGACAGGAGTGATCCTTAAAGATGTATCCTTCACGACACACAGCGGCGAAGTCACGGCCATACTGGGATCTAAAGGTAGCGGTAAACGCGCCCTCCTAGACGTGATAGCTCGTCGCGTGCCGTCAAAAGGCCACGTGCTACTCGAAGGCGTACCCTTAGAAGAGGAACAGTTCAACATCAACTGCGCTTTGGTTCGACACTCGACAAGACTGCTGCCAGGACTGAGCGTTCAGCAGACATTGGCCTTATCACTTACTAAGATTTCAGGGTATTTGAAGTCATCGAAAGTGAAACAAGTGATGGCTGACATGGCTCTCTCGCAGGTTGCGAACAAATGCGTCACAAATTTAACCAAAAGCGAATACAGAAGACTAGTTATTGGAGTGCAATTGATTCGTGATCCCATTATATTATTACTGGATGAGCCGACATGGGATTTGGATCCTCTCAACACATATCTTGTTATCTCAATATTATCGAATGCCGCTAAAAAATACGGGACCGCTATCATACTCACAATGGAAAAGCCTAGATCAGATGTATTCCCATTCCTGGACCGGGTGGTGTATCTATGCCTCGGGGATGTAGTATACGCGGGACCAACGAGGAACTTGCTTGACTACTTCGGCAGCATCGGGTTCCCGTGTCCACAGTTAGAAAACCCGCTCATGTACTACCTGTGCCTCTCCACCGTTGATAGGAGATCACGGGAGCGCTTCATTGAATCAAACCACCAGATCGCGGCGCTGGTCGAGAAATTTAAGATCGAAGGGCAAGCCATCATGCAGGGGAACCTTCATGAAAACAATCGTATCGTAAACCCTAATAAAGTACAAATGAACTATGGGAAGCCTGGCGGATTTAAGACAGTGTGGATGTTGTACTTACGGACTCTAGCTACAATATTCAATCTTAACAAACACGGATTCAAGCAAATGTTTATGAGACTTCTTTGTTTACCGATATATTTTTTCATACTTTGGATTTTCTACAACGAGGCAAAGGATTATCAACGGGCTTTCATTACTAAAAGCGGTTTAATCTTCAACGCAATGGTCGGAACTTATTTTGTCAGCATTATGAACACGATTTGTCTTTACGGTCCTTACAGAACTCGGTACTATGAAGAGTCCCAGGAGGGACATTACAGCGGCGCCAGTTTACTTCTGTCTTGGAATATAGTTTCGTTGCCATTTTCATGTATCACTTCGTTCGCTTCGGCTGCAATTATTTACCCGATCTTGGCCGACATTTCGGAATCGTTGGACTACATTCACTTCTCGTTGGTGCTGTGGTCGTCGTATATTTACGCGGAACAACAAGCGATCGCGATCATGGTATTCGTTAAGAACGGACTGAACGCCGCCATTGTCAACTTGTACATCACTTGCATTTACGTCATGCTCGCCAGTGGAGTGTTAAGATCCTACAAAGGCTTCGAAGACTGGCTCCACTACTTAACTTACATAACGCACACACGTTACGCCTCGATCTTCCTCCACCGGACCGTCTTCAAGCAACCGACCTTCAACATTCTGCCCTACAGCGACCACGAGAATTGCACGGGCATCACCAGCCTCATTCAGACCTCGTCGAGTTTAAACACGAATTCCAACGCTAACTGCAGATACGCCAGCGGGAAGGCATTCTTGACTGAACGATTCACGTCGAAAACTTTCACGGGGGACATTAACTTAGCGGGAGACTTCAATTTAAATTTCAATCTCGGCGTGTCGTTTGCATTCTCCGTCGGTATTATGATTTTCAACAAATTCCTGTATCTGATGCCTCTGCCTAGCTACATAACCGATAAATTCAGAGAATAA
Protein
MIGGDYTLELCNVFHSGQVEPGSFFQRLTGGVKTGVILKDVSFTTHSGEVTAILGSKGSGKRALLDVIARRVPSKGHVLLEGVPLEEEQFNINCALVRHSTRLLPGLSVQQTLALSLTKISGYLKSSKVKQVMADMALSQVANKCVTNLTKSEYRRLVIGVQLIRDPIILLLDEPTWDLDPLNTYLVISILSNAAKKYGTAIILTMEKPRSDVFPFLDRVVYLCLGDVVYAGPTRNLLDYFGSIGFPCPQLENPLMYYLCLSTVDRRSRERFIESNHQIAALVEKFKIEGQAIMQGNLHENNRIVNPNKVQMNYGKPGGFKTVWMLYLRTLATIFNLNKHGFKQMFMRLLCLPIYFFILWIFYNEAKDYQRAFITKSGLIFNAMVGTYFVSIMNTICLYGPYRTRYYEESQEGHYSGASLLLSWNIVSLPFSCITSFASAAIIYPILADISESLDYIHFSLVLWSSYIYAEQQAIAIMVFVKNGLNAAIVNLYITCIYVMLASGVLRSYKGFEDWLHYLTYITHTRYASIFLHRTVFKQPTFNILPYSDHENCTGITSLIQTSSSLNTNSNANCRYASGKAFLTERFTSKTFTGDINLAGDFNLNFNLGVSFAFSVGIMIFNKFLYLMPLPSYITDKFRE
Summary
Uniprot
A0A2A4JEL2
A0A2H1VEI0
A0A194Q7S4
A0A0N1IHT3
A0A3S2NKJ5
H9IT92
+ More
A0A212FCT4 A0A1Y1MH93 D6X0C7 A0A067RID4 A0A2J7R0T0 A0A1B6C953 U4TZU2 E0W1F2 J9JZK5 A0A1L3IWM2 A0A1C8C9L4 A0A1S3CYT8 A0A0K2T2T6 A0A0K2T3L7 A0A1I8NHC0 A0A2M4BHR3 A0A2P2I991 A0A340TBE5 A0A2M4CTI7 A0A1I8P7K3 A0A2M4A851 A0A2S2R741 A0A2C9GPN0 Q7PYP9 A0A1Y9H1Y7 A0A1A9XDK0 A0A0L0CQM0 B4M650 A0A336MS56 B4JYA7 A0A336M936 A0A0K8UQT0 A0A034VXW3 W8B4L0 A0A2H4TET1 B4KBS0 B4PV18 B3P6V9 A0A1W4VLI7 A0A1A9VJE7 B4G3K4 A0A0A1XT88 A0A1A9ZGE9 A0A1B0BTU2 Q29A05 B4NI47 A0A3B0JXU7 Q8MRJ2 B3LX06 B4HHN2 F4WRD6 A0A1B0G2E0 A0A154PBB4 A0A026W441 E9HF45 A0A151J2L3 A0A151I2I9 A0A158NUJ6 A0A195FK37 A0A0M4EJ10 E9IRV3 A0A146KX20 A0A224XLQ2 T1I8U5 E2C239 A0A069DW23 A0A087ZSN8 E2A381 A0A0P4W2N8 A0A0V0G7E6 A0A023F328 A0A0P5ZES6 A0A0N8DUZ2 A0A224YSS9 A0A131YU92 A0A2R5LFQ6 A0A0P5TDF2 A0A0P5KH12 A0A0P5IHX4 A0A0P5HJF3 A0A0L7QSM9 T1J9E6 A0A0P4ZH33 A0A087UTJ0 A0A2M3Z6E5 A0A1V9Y1Y3 A0A0P5DIK7 A0A195C7U0 A0A0P5AJI4 A0A1A9WRR3 A0A151WK50 A0A0P6HD43 A0A0N8ATA7
A0A212FCT4 A0A1Y1MH93 D6X0C7 A0A067RID4 A0A2J7R0T0 A0A1B6C953 U4TZU2 E0W1F2 J9JZK5 A0A1L3IWM2 A0A1C8C9L4 A0A1S3CYT8 A0A0K2T2T6 A0A0K2T3L7 A0A1I8NHC0 A0A2M4BHR3 A0A2P2I991 A0A340TBE5 A0A2M4CTI7 A0A1I8P7K3 A0A2M4A851 A0A2S2R741 A0A2C9GPN0 Q7PYP9 A0A1Y9H1Y7 A0A1A9XDK0 A0A0L0CQM0 B4M650 A0A336MS56 B4JYA7 A0A336M936 A0A0K8UQT0 A0A034VXW3 W8B4L0 A0A2H4TET1 B4KBS0 B4PV18 B3P6V9 A0A1W4VLI7 A0A1A9VJE7 B4G3K4 A0A0A1XT88 A0A1A9ZGE9 A0A1B0BTU2 Q29A05 B4NI47 A0A3B0JXU7 Q8MRJ2 B3LX06 B4HHN2 F4WRD6 A0A1B0G2E0 A0A154PBB4 A0A026W441 E9HF45 A0A151J2L3 A0A151I2I9 A0A158NUJ6 A0A195FK37 A0A0M4EJ10 E9IRV3 A0A146KX20 A0A224XLQ2 T1I8U5 E2C239 A0A069DW23 A0A087ZSN8 E2A381 A0A0P4W2N8 A0A0V0G7E6 A0A023F328 A0A0P5ZES6 A0A0N8DUZ2 A0A224YSS9 A0A131YU92 A0A2R5LFQ6 A0A0P5TDF2 A0A0P5KH12 A0A0P5IHX4 A0A0P5HJF3 A0A0L7QSM9 T1J9E6 A0A0P4ZH33 A0A087UTJ0 A0A2M3Z6E5 A0A1V9Y1Y3 A0A0P5DIK7 A0A195C7U0 A0A0P5AJI4 A0A1A9WRR3 A0A151WK50 A0A0P6HD43 A0A0N8ATA7
Pubmed
26354079
19121390
22118469
28004739
18362917
19820115
+ More
24845553 23537049 20566863 25315136 12364791 14747013 17210077 26108605 17994087 25348373 24495485 29121518 18057021 17550304 25830018 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21719571 24508170 30249741 21292972 21347285 21282665 26823975 20798317 26334808 27129103 25474469 30826642 28797301 26830274 28327890
24845553 23537049 20566863 25315136 12364791 14747013 17210077 26108605 17994087 25348373 24495485 29121518 18057021 17550304 25830018 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 21719571 24508170 30249741 21292972 21347285 21282665 26823975 20798317 26334808 27129103 25474469 30826642 28797301 26830274 28327890
EMBL
NWSH01001872
PCG69842.1
ODYU01002136
SOQ39243.1
KQ459324
KPJ01587.1
+ More
KQ460041 KPJ18364.1 RSAL01000354 RVE42240.1 BABH01013556 AGBW02009152 OWR51551.1 GEZM01034552 JAV83735.1 KQ971372 EFA10526.2 KK852616 KDR20187.1 NEVH01008218 PNF34425.1 GEDC01027439 GEDC01018128 GEDC01012799 JAS09859.1 JAS19170.1 JAS24499.1 KB631924 ERL87154.1 DS235870 EEB19458.1 ABLF02027753 ABLF02027757 KX056442 APG57152.1 KJ863734 AIO05330.1 MH172518 QAA95925.1 HACA01003008 CDW20369.1 HACA01003009 CDW20370.1 GGFJ01003455 MBW52596.1 IACF01004899 LAB70486.1 GGFL01004411 MBW68589.1 GGFK01003655 MBW36976.1 GGMS01016628 MBY85831.1 APCN01002031 AAAB01008987 EAA01062.5 JRES01000062 KNC34477.1 CH940652 EDW59126.2 UFQS01002049 UFQT01002049 SSX13095.1 SSX32535.1 CH916377 EDV90669.1 UFQT01000575 SSX25429.1 GDHF01023272 JAI29042.1 GAKP01012272 GAKP01012271 JAC46680.1 GAMC01014567 JAB91988.1 KY849644 ATY74525.1 CH933806 EDW14747.1 KRG01177.1 CM000160 EDW98867.1 KRK04507.1 CH954182 EDV53779.1 CH479179 EDW25012.1 GBXI01000112 JAD14180.1 JXJN01020356 JXJN01020357 CM000070 EAL27546.2 CH964272 EDW84739.1 OUUW01000005 SPP80330.1 AE014297 AY119582 AAF56361.2 AAM50236.1 CH902617 EDV43845.1 CH480815 EDW43574.1 GL888285 EGI63207.1 CCAG010007069 KQ434868 KZC09185.1 KK107496 QOIP01000004 EZA49814.1 RLU24091.1 GL732633 EFX69652.1 KQ980368 KYN16328.1 KQ976533 KYM81498.1 ADTU01026585 KQ981522 KYN40627.1 CP012526 ALC46784.1 GL765283 EFZ16691.1 GDHC01017616 JAQ01013.1 GFTR01007475 JAW08951.1 ACPB03001910 GL452067 EFN77990.1 GBGD01000848 JAC88041.1 GL436311 EFN72109.1 GDKW01000820 JAI55775.1 GECL01002861 JAP03263.1 GBBI01003191 JAC15521.1 GDIP01044704 LRGB01003375 MK520846 JAM59011.1 KZS03020.1 QBM06379.1 GDIQ01089332 JAN05405.1 GFPF01006197 MAA17343.1 GEDV01006020 JAP82537.1 GGLE01004173 MBY08299.1 GDIP01133669 JAL70045.1 GDIQ01189601 JAK62124.1 GDIQ01237950 JAK13775.1 GDIQ01227238 JAK24487.1 KQ414756 KOC61647.1 JH431972 GDIP01213560 JAJ09842.1 KK121536 KFM80679.1 GGFM01003348 MBW24099.1 MNPL01000773 OQR79715.1 GDIP01156739 JAJ66663.1 KQ978143 KYM96715.1 GDIP01197882 JAJ25520.1 KQ983014 KYQ48242.1 GDIQ01021089 JAN73648.1 GDIQ01245003 JAK06722.1
KQ460041 KPJ18364.1 RSAL01000354 RVE42240.1 BABH01013556 AGBW02009152 OWR51551.1 GEZM01034552 JAV83735.1 KQ971372 EFA10526.2 KK852616 KDR20187.1 NEVH01008218 PNF34425.1 GEDC01027439 GEDC01018128 GEDC01012799 JAS09859.1 JAS19170.1 JAS24499.1 KB631924 ERL87154.1 DS235870 EEB19458.1 ABLF02027753 ABLF02027757 KX056442 APG57152.1 KJ863734 AIO05330.1 MH172518 QAA95925.1 HACA01003008 CDW20369.1 HACA01003009 CDW20370.1 GGFJ01003455 MBW52596.1 IACF01004899 LAB70486.1 GGFL01004411 MBW68589.1 GGFK01003655 MBW36976.1 GGMS01016628 MBY85831.1 APCN01002031 AAAB01008987 EAA01062.5 JRES01000062 KNC34477.1 CH940652 EDW59126.2 UFQS01002049 UFQT01002049 SSX13095.1 SSX32535.1 CH916377 EDV90669.1 UFQT01000575 SSX25429.1 GDHF01023272 JAI29042.1 GAKP01012272 GAKP01012271 JAC46680.1 GAMC01014567 JAB91988.1 KY849644 ATY74525.1 CH933806 EDW14747.1 KRG01177.1 CM000160 EDW98867.1 KRK04507.1 CH954182 EDV53779.1 CH479179 EDW25012.1 GBXI01000112 JAD14180.1 JXJN01020356 JXJN01020357 CM000070 EAL27546.2 CH964272 EDW84739.1 OUUW01000005 SPP80330.1 AE014297 AY119582 AAF56361.2 AAM50236.1 CH902617 EDV43845.1 CH480815 EDW43574.1 GL888285 EGI63207.1 CCAG010007069 KQ434868 KZC09185.1 KK107496 QOIP01000004 EZA49814.1 RLU24091.1 GL732633 EFX69652.1 KQ980368 KYN16328.1 KQ976533 KYM81498.1 ADTU01026585 KQ981522 KYN40627.1 CP012526 ALC46784.1 GL765283 EFZ16691.1 GDHC01017616 JAQ01013.1 GFTR01007475 JAW08951.1 ACPB03001910 GL452067 EFN77990.1 GBGD01000848 JAC88041.1 GL436311 EFN72109.1 GDKW01000820 JAI55775.1 GECL01002861 JAP03263.1 GBBI01003191 JAC15521.1 GDIP01044704 LRGB01003375 MK520846 JAM59011.1 KZS03020.1 QBM06379.1 GDIQ01089332 JAN05405.1 GFPF01006197 MAA17343.1 GEDV01006020 JAP82537.1 GGLE01004173 MBY08299.1 GDIP01133669 JAL70045.1 GDIQ01189601 JAK62124.1 GDIQ01237950 JAK13775.1 GDIQ01227238 JAK24487.1 KQ414756 KOC61647.1 JH431972 GDIP01213560 JAJ09842.1 KK121536 KFM80679.1 GGFM01003348 MBW24099.1 MNPL01000773 OQR79715.1 GDIP01156739 JAJ66663.1 KQ978143 KYM96715.1 GDIP01197882 JAJ25520.1 KQ983014 KYQ48242.1 GDIQ01021089 JAN73648.1 GDIQ01245003 JAK06722.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000283053
UP000005204
UP000007151
+ More
UP000007266 UP000027135 UP000235965 UP000030742 UP000009046 UP000007819 UP000079169 UP000095301 UP000076407 UP000095300 UP000075840 UP000007062 UP000075884 UP000092443 UP000037069 UP000008792 UP000001070 UP000009192 UP000002282 UP000008711 UP000192221 UP000078200 UP000008744 UP000092445 UP000092460 UP000001819 UP000007798 UP000268350 UP000000803 UP000007801 UP000001292 UP000007755 UP000092444 UP000076502 UP000053097 UP000279307 UP000000305 UP000078492 UP000078540 UP000005205 UP000078541 UP000092553 UP000015103 UP000008237 UP000005203 UP000000311 UP000076858 UP000053825 UP000054359 UP000192247 UP000078542 UP000091820 UP000075809
UP000007266 UP000027135 UP000235965 UP000030742 UP000009046 UP000007819 UP000079169 UP000095301 UP000076407 UP000095300 UP000075840 UP000007062 UP000075884 UP000092443 UP000037069 UP000008792 UP000001070 UP000009192 UP000002282 UP000008711 UP000192221 UP000078200 UP000008744 UP000092445 UP000092460 UP000001819 UP000007798 UP000268350 UP000000803 UP000007801 UP000001292 UP000007755 UP000092444 UP000076502 UP000053097 UP000279307 UP000000305 UP000078492 UP000078540 UP000005205 UP000078541 UP000092553 UP000015103 UP000008237 UP000005203 UP000000311 UP000076858 UP000053825 UP000054359 UP000192247 UP000078542 UP000091820 UP000075809
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2A4JEL2
A0A2H1VEI0
A0A194Q7S4
A0A0N1IHT3
A0A3S2NKJ5
H9IT92
+ More
A0A212FCT4 A0A1Y1MH93 D6X0C7 A0A067RID4 A0A2J7R0T0 A0A1B6C953 U4TZU2 E0W1F2 J9JZK5 A0A1L3IWM2 A0A1C8C9L4 A0A1S3CYT8 A0A0K2T2T6 A0A0K2T3L7 A0A1I8NHC0 A0A2M4BHR3 A0A2P2I991 A0A340TBE5 A0A2M4CTI7 A0A1I8P7K3 A0A2M4A851 A0A2S2R741 A0A2C9GPN0 Q7PYP9 A0A1Y9H1Y7 A0A1A9XDK0 A0A0L0CQM0 B4M650 A0A336MS56 B4JYA7 A0A336M936 A0A0K8UQT0 A0A034VXW3 W8B4L0 A0A2H4TET1 B4KBS0 B4PV18 B3P6V9 A0A1W4VLI7 A0A1A9VJE7 B4G3K4 A0A0A1XT88 A0A1A9ZGE9 A0A1B0BTU2 Q29A05 B4NI47 A0A3B0JXU7 Q8MRJ2 B3LX06 B4HHN2 F4WRD6 A0A1B0G2E0 A0A154PBB4 A0A026W441 E9HF45 A0A151J2L3 A0A151I2I9 A0A158NUJ6 A0A195FK37 A0A0M4EJ10 E9IRV3 A0A146KX20 A0A224XLQ2 T1I8U5 E2C239 A0A069DW23 A0A087ZSN8 E2A381 A0A0P4W2N8 A0A0V0G7E6 A0A023F328 A0A0P5ZES6 A0A0N8DUZ2 A0A224YSS9 A0A131YU92 A0A2R5LFQ6 A0A0P5TDF2 A0A0P5KH12 A0A0P5IHX4 A0A0P5HJF3 A0A0L7QSM9 T1J9E6 A0A0P4ZH33 A0A087UTJ0 A0A2M3Z6E5 A0A1V9Y1Y3 A0A0P5DIK7 A0A195C7U0 A0A0P5AJI4 A0A1A9WRR3 A0A151WK50 A0A0P6HD43 A0A0N8ATA7
A0A212FCT4 A0A1Y1MH93 D6X0C7 A0A067RID4 A0A2J7R0T0 A0A1B6C953 U4TZU2 E0W1F2 J9JZK5 A0A1L3IWM2 A0A1C8C9L4 A0A1S3CYT8 A0A0K2T2T6 A0A0K2T3L7 A0A1I8NHC0 A0A2M4BHR3 A0A2P2I991 A0A340TBE5 A0A2M4CTI7 A0A1I8P7K3 A0A2M4A851 A0A2S2R741 A0A2C9GPN0 Q7PYP9 A0A1Y9H1Y7 A0A1A9XDK0 A0A0L0CQM0 B4M650 A0A336MS56 B4JYA7 A0A336M936 A0A0K8UQT0 A0A034VXW3 W8B4L0 A0A2H4TET1 B4KBS0 B4PV18 B3P6V9 A0A1W4VLI7 A0A1A9VJE7 B4G3K4 A0A0A1XT88 A0A1A9ZGE9 A0A1B0BTU2 Q29A05 B4NI47 A0A3B0JXU7 Q8MRJ2 B3LX06 B4HHN2 F4WRD6 A0A1B0G2E0 A0A154PBB4 A0A026W441 E9HF45 A0A151J2L3 A0A151I2I9 A0A158NUJ6 A0A195FK37 A0A0M4EJ10 E9IRV3 A0A146KX20 A0A224XLQ2 T1I8U5 E2C239 A0A069DW23 A0A087ZSN8 E2A381 A0A0P4W2N8 A0A0V0G7E6 A0A023F328 A0A0P5ZES6 A0A0N8DUZ2 A0A224YSS9 A0A131YU92 A0A2R5LFQ6 A0A0P5TDF2 A0A0P5KH12 A0A0P5IHX4 A0A0P5HJF3 A0A0L7QSM9 T1J9E6 A0A0P4ZH33 A0A087UTJ0 A0A2M3Z6E5 A0A1V9Y1Y3 A0A0P5DIK7 A0A195C7U0 A0A0P5AJI4 A0A1A9WRR3 A0A151WK50 A0A0P6HD43 A0A0N8ATA7
PDB
5DO7
E-value=3.62151e-54,
Score=537
Ontologies
GO
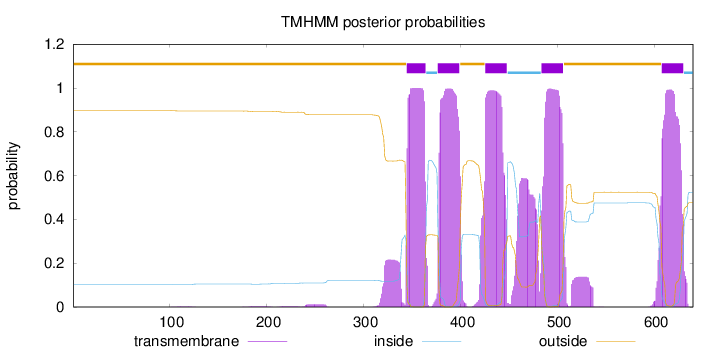
Topology
Length:
640
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
128.83893
Exp number, first 60 AAs:
0.00185
Total prob of N-in:
0.10318
outside
1 - 344
TMhelix
345 - 364
inside
365 - 376
TMhelix
377 - 399
outside
400 - 425
TMhelix
426 - 448
inside
449 - 483
TMhelix
484 - 506
outside
507 - 607
TMhelix
608 - 630
inside
631 - 640
Population Genetic Test Statistics
Pi
251.746143
Theta
154.177646
Tajima's D
1.629775
CLR
0.331638
CSRT
0.810659467026649
Interpretation
Uncertain
