Pre Gene Modal
BGIBMGA012690
Annotation
PREDICTED:_pre-rRNA-processing_protein_TSR1_homolog_[Amyelois_transitella]
Full name
Pre-rRNA-processing protein TSR1 homolog
Location in the cell
Nuclear Reliability : 2.816
Sequence
CDS
ATGCAACAACCACATCGTCCCGGCCATTTCAAGCAGAGCAATAAAACTCATAAATCACGCCATCGCTCAAAACGAGGTATCGCCGCTGCTGTAAAAGGAAAAGTCAACTTGAAAGAAATTGTTCGACGTAATCGTCGTGAGCTTAGAAAAGACGATCGCCGTCATCAGGCTTCACAAATTCGCAAAAATAAAAGGGAAGAGGTTCTAGCTACGAAACGTGCTTTAGGAGGCACTAGGAATCCGCCGTTTCTAGTTTGTGTCGTTCCGCTTAATGCGCAACTCGATGTAGGATCGGCACTCGCCATTCTGAAGACCTGTTCTGATGGGGCTGTCGTAAACGAGTCTGAAAACGGAATTGTACACATCGGGTTGCCAAATTTCAAACAGCGGTTTTCGTTTATTTGTCCTGAAGTAGGTGACGACTTTGCACTTCTTGATGTTTTAAAAATAGCTGACACTGCCCTCTATGTAAGCTCAGCTTTAGAGGAACCAGTTGATGAATGGGGAGAAAAAGTTTTAGCACTCTCTATGGCCCAAGGAATGCCTACCCCTGTAGTAGTAGCAATGGATATAGAGGGAGTCCACCCAAAGAAACGCACTACAGAAAAACAGAATGTTCAGAAATTGGTCTCAAAATGGCTTCCAGAAGAAAAGGTAATGCAATTAGATAAAAAATCAGATGGTTTGAACTTGTTAAGGAAAATAGGAAATCAGAAACGTAACATAATACATCATAGAGAGAAAAGACCTTATATGCTGGCTGAGGAAGTTGAATATGTACCCGATTTAGAAGGTAACAGTGGTACTTTAAGAGTATCAGGGTATTTACGAGGAATGGCACTTAATGTCAATGGACTTGTTCATATTACTGGACTTGGTGACTTTCAAATGACTAGAATAGAGGGTTTTGAGGACCCCCACCCATTTCATTTGGGGAAAGAAAATACTAATGATGATGGTATGTATGCTGAAGTCACAAAAATGTCAGTTCTCGCAGTAGCTGATCCTGCGAAGCAAGAGAGTCTTCAATCAGAGAATGTGCCAGACCCTATGGATGCAGAGCAGACCTGGCCAACTGAAGAAGAAATTGAACAAGCTAATTTAGAGACACAAAAGAAAAAAATCAAGAAAGTACCCAAAGGTTGGTCTGATTACCAAGCTGCTTGGATTGTCGAATCTGATGCCGAGGAAGATGGTGATGAGTCTGATAGTGGAGATGAGAAAGAAAACGATGAGTTTATGTCTTGTGAAGATGATAATTCTGATCCAGAGGACGATAAACAGTTCAATGATATTGAATCTGTTACCGAATCAGAGGTTGGACCCACAGATGAGAAGTATGATGCCACAATAGACACTCAGGAAGAACATGAGATGTTGCAGAAGTTAGCAGCTGCAAAAGAAGATCAGCAATTCCCTGATGAAGTTGATACCCCGCAAGATATCCCAGCTAGAGAGCGTTTTATGAGATACAGAGGACTAGAGTCTTTCAGGACATCAGCATGGGATGTAAAGGAAAATTTGCCTCAGGATTATTCTAGAATATTTCAGTTTGAAAACTATGAGCGCACCAGGAAGAGGGTTTTTAAGGAACTCGAAGACAGTTTAGTTAATATGTATGGCTTTTATATAAGAATACATGTAAAAGGTGTTCATCAAGATTTATGGAAAGCCTTTAGTACAGCAAATCCGAATGCTCCGTTGGCTGTTTTCGGACTTTTACCACACGAACACAAAATGTCTCTGATGAATGTAGTATTAAAACGCACTGGTGCCTGCAATGAACCTATCAAGAGCAAGGAAAGACTTATATTCCAAGTTGGTTATAGAAGGTTCATTGTGAATCCTATATTCAGTCAGCACACCAATGGCAGTAAGCATAAGTACGAGAGATTCTTCCAACCAGCATCAACTTGTGTTGCATCTTTCTTTGCACCTATACAGTTCAGTCCATCTACAGTGCTATGCTTTAAGGAAAAAAAGAACACAAAGCTGCAGCTTGTAGCAACTGGCGTACTACTCTCATGCAGCCCTGATAGATTAGTTATAAAAAGAATTGTACTCTCTGGACATCCATATAAGGTTCATAAAAAGTCAGCCGTCATAAGATTCATGTTCTTTAACAGAGAAGATATAGTTTACTTCAAGCCCTGTAAACTAAGATCGAAGTATGGGCGTACTGGACACATCAAAGAACCTTTGGGTACTCACGGTCATATGAAATGCGTTTTCGACGGGCAACTGAAATCACAAGATACCGTACTATTAAACTTATACAAACGAATCTTCCCGAAATGGTCCTACGAGAACTGTATAGTTACAAGCAAAGACAATGATGTTGTAATGGAGTAA
Protein
MQQPHRPGHFKQSNKTHKSRHRSKRGIAAAVKGKVNLKEIVRRNRRELRKDDRRHQASQIRKNKREEVLATKRALGGTRNPPFLVCVVPLNAQLDVGSALAILKTCSDGAVVNESENGIVHIGLPNFKQRFSFICPEVGDDFALLDVLKIADTALYVSSALEEPVDEWGEKVLALSMAQGMPTPVVVAMDIEGVHPKKRTTEKQNVQKLVSKWLPEEKVMQLDKKSDGLNLLRKIGNQKRNIIHHREKRPYMLAEEVEYVPDLEGNSGTLRVSGYLRGMALNVNGLVHITGLGDFQMTRIEGFEDPHPFHLGKENTNDDGMYAEVTKMSVLAVADPAKQESLQSENVPDPMDAEQTWPTEEEIEQANLETQKKKIKKVPKGWSDYQAAWIVESDAEEDGDESDSGDEKENDEFMSCEDDNSDPEDDKQFNDIESVTESEVGPTDEKYDATIDTQEEHEMLQKLAAAKEDQQFPDEVDTPQDIPARERFMRYRGLESFRTSAWDVKENLPQDYSRIFQFENYERTRKRVFKELEDSLVNMYGFYIRIHVKGVHQDLWKAFSTANPNAPLAVFGLLPHEHKMSLMNVVLKRTGACNEPIKSKERLIFQVGYRRFIVNPIFSQHTNGSKHKYERFFQPASTCVASFFAPIQFSPSTVLCFKEKKNTKLQLVATGVLLSCSPDRLVIKRIVLSGHPYKVHKKSAVIRFMFFNREDIVYFKPCKLRSKYGRTGHIKEPLGTHGHMKCVFDGQLKSQDTVLLNLYKRIFPKWSYENCIVTSKDNDVVME
Summary
Description
Required during maturation of the 40S ribosomal subunit in the nucleolus.
Similarity
Belongs to the TRAFAC class translation factor GTPase superfamily. Bms1-like GTPase family. TSR1 subfamily.
Keywords
Complete proteome
Nucleus
Phosphoprotein
Reference proteome
Ribosome biogenesis
Feature
chain Pre-rRNA-processing protein TSR1 homolog
Uniprot
H9JT28
A0A2H1UZY8
A0A2A4JCW0
S4PLJ2
A0A212F4Y4
A0A0N1IJ17
+ More
A0A194Q7T8 A0A2W1BG93 D7EJ12 A0A1Y1NAY7 A0A182TLU0 A0A182XIN5 Q0IFN5 A0A182L5E8 A0A0K8TNK8 A0A182HG52 Q7PTN1 A0A182VUS9 A0A182YFY5 B4KWV0 A0A182VBZ3 A0A182QZG5 A0A1Q3F227 A0A1Q3F1T9 U5EVS0 A0A182K2L8 A0A1S4F7T6 A0A1Q3F234 A0A182PJ12 A0A182JI51 B4LH21 A0A087ZSW0 A0A182G912 A0A182M2Y5 A0A182RG97 B0W236 A0A182GYZ8 T1PFG7 A0A0L7RA28 A0A1W4XNG4 A0A182F5B1 A0A0L0BVP6 A0A2A3EDS7 A0A310SHZ3 A0A084VPS7 A0A1W4VAH3 A0A0J9S0Q0 A0A182NEL4 A0A2M3Z0Z7 B4QJR6 B4PE42 B4PE37 A0A0M8ZX74 B4N9X1 A0A2M3Z0X1 A0A1I8PFG8 B4IAN4 B3NE72 B4J236 W5J905 T1DGP0 A0A0M4EAD5 A0A2M4BDS3 A0A2M4A6D1 A0A2M4BDS8 A0A1A9X530 Q9VP47 B4H605 K7IVL4 A0A2M4A635 A0A3B0KNC2 B3M8I4 Q2LYP0 A0A2M4A8R5 A0A2M4BDX3 A0A232F751 A0A1A9VYH2 A0A0K8V8L3 A0A067RP15 A0A0A1WIV2 A0A1A9Y9J4 W8C398 A0A154PB96 A0A1B0B9E2 A0A1J1J3R8 A0A1A9Z3I8 U4UNY1 A0A1B0FC46 N6ULQ5 A0A151WFB6 A0A195F2S0 F4WM77 A0A195DDQ4 A0A0L7L9J7 A0A034VY99 A0A195BD14 E9IEX7 A0A158P334 A0A151IGD8 A0A1Y1N7A1
A0A194Q7T8 A0A2W1BG93 D7EJ12 A0A1Y1NAY7 A0A182TLU0 A0A182XIN5 Q0IFN5 A0A182L5E8 A0A0K8TNK8 A0A182HG52 Q7PTN1 A0A182VUS9 A0A182YFY5 B4KWV0 A0A182VBZ3 A0A182QZG5 A0A1Q3F227 A0A1Q3F1T9 U5EVS0 A0A182K2L8 A0A1S4F7T6 A0A1Q3F234 A0A182PJ12 A0A182JI51 B4LH21 A0A087ZSW0 A0A182G912 A0A182M2Y5 A0A182RG97 B0W236 A0A182GYZ8 T1PFG7 A0A0L7RA28 A0A1W4XNG4 A0A182F5B1 A0A0L0BVP6 A0A2A3EDS7 A0A310SHZ3 A0A084VPS7 A0A1W4VAH3 A0A0J9S0Q0 A0A182NEL4 A0A2M3Z0Z7 B4QJR6 B4PE42 B4PE37 A0A0M8ZX74 B4N9X1 A0A2M3Z0X1 A0A1I8PFG8 B4IAN4 B3NE72 B4J236 W5J905 T1DGP0 A0A0M4EAD5 A0A2M4BDS3 A0A2M4A6D1 A0A2M4BDS8 A0A1A9X530 Q9VP47 B4H605 K7IVL4 A0A2M4A635 A0A3B0KNC2 B3M8I4 Q2LYP0 A0A2M4A8R5 A0A2M4BDX3 A0A232F751 A0A1A9VYH2 A0A0K8V8L3 A0A067RP15 A0A0A1WIV2 A0A1A9Y9J4 W8C398 A0A154PB96 A0A1B0B9E2 A0A1J1J3R8 A0A1A9Z3I8 U4UNY1 A0A1B0FC46 N6ULQ5 A0A151WFB6 A0A195F2S0 F4WM77 A0A195DDQ4 A0A0L7L9J7 A0A034VY99 A0A195BD14 E9IEX7 A0A158P334 A0A151IGD8 A0A1Y1N7A1
Pubmed
19121390
23622113
22118469
26354079
28756777
18362917
+ More
19820115 28004739 17510324 20966253 26369729 12364791 14747013 17210077 25244985 17994087 26483478 25315136 26108605 24438588 22936249 17550304 20920257 23761445 10731132 12537572 12537569 18327897 20075255 15632085 28648823 24845553 25830018 24495485 23537049 21719571 26227816 25348373 21282665 21347285
19820115 28004739 17510324 20966253 26369729 12364791 14747013 17210077 25244985 17994087 26483478 25315136 26108605 24438588 22936249 17550304 20920257 23761445 10731132 12537572 12537569 18327897 20075255 15632085 28648823 24845553 25830018 24495485 23537049 21719571 26227816 25348373 21282665 21347285
EMBL
BABH01030658
ODYU01000045
SOQ34151.1
NWSH01002087
PCG69233.1
GAIX01004000
+ More
JAA88560.1 AGBW02010296 OWR48790.1 KQ460041 KPJ18350.1 KQ459324 KPJ01602.1 KZ150492 PZC70683.1 DS497692 EFA12491.1 GEZM01010819 JAV93805.1 CH477306 EAT44108.1 GDAI01001902 JAI15701.1 APCN01004115 AAAB01008797 EAA03633.5 CH933809 EDW18571.1 AXCN02000850 GFDL01013435 JAV21610.1 GFDL01013521 JAV21524.1 GANO01001743 JAB58128.1 GFDL01013429 JAV21616.1 CH940647 EDW69511.1 JXUM01048556 KQ561567 KXJ78145.1 AXCM01003253 DS231825 EDS28164.1 JXUM01098527 KQ564427 KXJ72297.1 KA646653 AFP61282.1 KQ414619 KOC67732.1 JRES01001262 KNC24107.1 KZ288280 PBC29634.1 KQ760974 OAD58581.1 ATLV01015034 KE524999 KFB39971.1 CM002912 KMZ00980.1 GGFM01001448 MBW22199.1 CM000363 EDX11366.1 CM000159 EDW95055.1 EDW95050.1 KQ435810 KOX72945.1 CH964232 EDW81726.1 GGFM01001413 MBW22164.1 CH480826 EDW44347.1 CH954178 EDV52636.1 CH916366 EDV96987.1 ADMH02002093 ETN59284.1 GAMD01002701 JAA98889.1 CP012525 ALC44204.1 GGFJ01002051 MBW51192.1 GGFK01002994 MBW36315.1 GGFJ01002053 MBW51194.1 AE014296 BT003192 BT044092 AY058591 CH479212 EDW33222.1 GGFK01002861 MBW36182.1 OUUW01000009 SPP85318.1 CH902618 EDV39958.1 CH379069 EAL29821.2 GGFK01003687 MBW37008.1 GGFJ01002052 MBW51193.1 NNAY01000752 OXU26684.1 GDHF01017050 JAI35264.1 KK852510 KDR22360.1 GBXI01015515 JAC98776.1 GAMC01000128 JAC06428.1 KQ434868 KZC09196.1 JXJN01010368 CVRI01000070 CRL07083.1 KB632303 ERL91831.1 CCAG010016063 APGK01018922 KB740085 ENN81566.1 KQ983219 KYQ46531.1 KQ981855 KYN34758.1 GL888217 EGI64721.1 KQ980989 KYN10559.1 JTDY01002117 KOB72070.1 GAKP01011543 JAC47409.1 KQ976522 KYM82080.1 GL762728 EFZ20870.1 ADTU01001302 KQ977726 KYN00287.1 GEZM01010820 GEZM01010818 JAV93804.1
JAA88560.1 AGBW02010296 OWR48790.1 KQ460041 KPJ18350.1 KQ459324 KPJ01602.1 KZ150492 PZC70683.1 DS497692 EFA12491.1 GEZM01010819 JAV93805.1 CH477306 EAT44108.1 GDAI01001902 JAI15701.1 APCN01004115 AAAB01008797 EAA03633.5 CH933809 EDW18571.1 AXCN02000850 GFDL01013435 JAV21610.1 GFDL01013521 JAV21524.1 GANO01001743 JAB58128.1 GFDL01013429 JAV21616.1 CH940647 EDW69511.1 JXUM01048556 KQ561567 KXJ78145.1 AXCM01003253 DS231825 EDS28164.1 JXUM01098527 KQ564427 KXJ72297.1 KA646653 AFP61282.1 KQ414619 KOC67732.1 JRES01001262 KNC24107.1 KZ288280 PBC29634.1 KQ760974 OAD58581.1 ATLV01015034 KE524999 KFB39971.1 CM002912 KMZ00980.1 GGFM01001448 MBW22199.1 CM000363 EDX11366.1 CM000159 EDW95055.1 EDW95050.1 KQ435810 KOX72945.1 CH964232 EDW81726.1 GGFM01001413 MBW22164.1 CH480826 EDW44347.1 CH954178 EDV52636.1 CH916366 EDV96987.1 ADMH02002093 ETN59284.1 GAMD01002701 JAA98889.1 CP012525 ALC44204.1 GGFJ01002051 MBW51192.1 GGFK01002994 MBW36315.1 GGFJ01002053 MBW51194.1 AE014296 BT003192 BT044092 AY058591 CH479212 EDW33222.1 GGFK01002861 MBW36182.1 OUUW01000009 SPP85318.1 CH902618 EDV39958.1 CH379069 EAL29821.2 GGFK01003687 MBW37008.1 GGFJ01002052 MBW51193.1 NNAY01000752 OXU26684.1 GDHF01017050 JAI35264.1 KK852510 KDR22360.1 GBXI01015515 JAC98776.1 GAMC01000128 JAC06428.1 KQ434868 KZC09196.1 JXJN01010368 CVRI01000070 CRL07083.1 KB632303 ERL91831.1 CCAG010016063 APGK01018922 KB740085 ENN81566.1 KQ983219 KYQ46531.1 KQ981855 KYN34758.1 GL888217 EGI64721.1 KQ980989 KYN10559.1 JTDY01002117 KOB72070.1 GAKP01011543 JAC47409.1 KQ976522 KYM82080.1 GL762728 EFZ20870.1 ADTU01001302 KQ977726 KYN00287.1 GEZM01010820 GEZM01010818 JAV93804.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000007266
+ More
UP000075902 UP000076407 UP000008820 UP000075882 UP000075840 UP000007062 UP000075920 UP000076408 UP000009192 UP000075903 UP000075886 UP000075881 UP000075885 UP000075880 UP000008792 UP000005203 UP000069940 UP000249989 UP000075883 UP000075900 UP000002320 UP000095301 UP000053825 UP000192223 UP000069272 UP000037069 UP000242457 UP000030765 UP000192221 UP000075884 UP000000304 UP000002282 UP000053105 UP000007798 UP000095300 UP000001292 UP000008711 UP000001070 UP000000673 UP000092553 UP000091820 UP000000803 UP000008744 UP000002358 UP000268350 UP000007801 UP000001819 UP000215335 UP000078200 UP000027135 UP000092443 UP000076502 UP000092460 UP000183832 UP000092445 UP000030742 UP000092444 UP000019118 UP000075809 UP000078541 UP000007755 UP000078492 UP000037510 UP000078540 UP000005205 UP000078542
UP000075902 UP000076407 UP000008820 UP000075882 UP000075840 UP000007062 UP000075920 UP000076408 UP000009192 UP000075903 UP000075886 UP000075881 UP000075885 UP000075880 UP000008792 UP000005203 UP000069940 UP000249989 UP000075883 UP000075900 UP000002320 UP000095301 UP000053825 UP000192223 UP000069272 UP000037069 UP000242457 UP000030765 UP000192221 UP000075884 UP000000304 UP000002282 UP000053105 UP000007798 UP000095300 UP000001292 UP000008711 UP000001070 UP000000673 UP000092553 UP000091820 UP000000803 UP000008744 UP000002358 UP000268350 UP000007801 UP000001819 UP000215335 UP000078200 UP000027135 UP000092443 UP000076502 UP000092460 UP000183832 UP000092445 UP000030742 UP000092444 UP000019118 UP000075809 UP000078541 UP000007755 UP000078492 UP000037510 UP000078540 UP000005205 UP000078542
Interpro
SUPFAM
SSF48371
SSF48371
ProteinModelPortal
H9JT28
A0A2H1UZY8
A0A2A4JCW0
S4PLJ2
A0A212F4Y4
A0A0N1IJ17
+ More
A0A194Q7T8 A0A2W1BG93 D7EJ12 A0A1Y1NAY7 A0A182TLU0 A0A182XIN5 Q0IFN5 A0A182L5E8 A0A0K8TNK8 A0A182HG52 Q7PTN1 A0A182VUS9 A0A182YFY5 B4KWV0 A0A182VBZ3 A0A182QZG5 A0A1Q3F227 A0A1Q3F1T9 U5EVS0 A0A182K2L8 A0A1S4F7T6 A0A1Q3F234 A0A182PJ12 A0A182JI51 B4LH21 A0A087ZSW0 A0A182G912 A0A182M2Y5 A0A182RG97 B0W236 A0A182GYZ8 T1PFG7 A0A0L7RA28 A0A1W4XNG4 A0A182F5B1 A0A0L0BVP6 A0A2A3EDS7 A0A310SHZ3 A0A084VPS7 A0A1W4VAH3 A0A0J9S0Q0 A0A182NEL4 A0A2M3Z0Z7 B4QJR6 B4PE42 B4PE37 A0A0M8ZX74 B4N9X1 A0A2M3Z0X1 A0A1I8PFG8 B4IAN4 B3NE72 B4J236 W5J905 T1DGP0 A0A0M4EAD5 A0A2M4BDS3 A0A2M4A6D1 A0A2M4BDS8 A0A1A9X530 Q9VP47 B4H605 K7IVL4 A0A2M4A635 A0A3B0KNC2 B3M8I4 Q2LYP0 A0A2M4A8R5 A0A2M4BDX3 A0A232F751 A0A1A9VYH2 A0A0K8V8L3 A0A067RP15 A0A0A1WIV2 A0A1A9Y9J4 W8C398 A0A154PB96 A0A1B0B9E2 A0A1J1J3R8 A0A1A9Z3I8 U4UNY1 A0A1B0FC46 N6ULQ5 A0A151WFB6 A0A195F2S0 F4WM77 A0A195DDQ4 A0A0L7L9J7 A0A034VY99 A0A195BD14 E9IEX7 A0A158P334 A0A151IGD8 A0A1Y1N7A1
A0A194Q7T8 A0A2W1BG93 D7EJ12 A0A1Y1NAY7 A0A182TLU0 A0A182XIN5 Q0IFN5 A0A182L5E8 A0A0K8TNK8 A0A182HG52 Q7PTN1 A0A182VUS9 A0A182YFY5 B4KWV0 A0A182VBZ3 A0A182QZG5 A0A1Q3F227 A0A1Q3F1T9 U5EVS0 A0A182K2L8 A0A1S4F7T6 A0A1Q3F234 A0A182PJ12 A0A182JI51 B4LH21 A0A087ZSW0 A0A182G912 A0A182M2Y5 A0A182RG97 B0W236 A0A182GYZ8 T1PFG7 A0A0L7RA28 A0A1W4XNG4 A0A182F5B1 A0A0L0BVP6 A0A2A3EDS7 A0A310SHZ3 A0A084VPS7 A0A1W4VAH3 A0A0J9S0Q0 A0A182NEL4 A0A2M3Z0Z7 B4QJR6 B4PE42 B4PE37 A0A0M8ZX74 B4N9X1 A0A2M3Z0X1 A0A1I8PFG8 B4IAN4 B3NE72 B4J236 W5J905 T1DGP0 A0A0M4EAD5 A0A2M4BDS3 A0A2M4A6D1 A0A2M4BDS8 A0A1A9X530 Q9VP47 B4H605 K7IVL4 A0A2M4A635 A0A3B0KNC2 B3M8I4 Q2LYP0 A0A2M4A8R5 A0A2M4BDX3 A0A232F751 A0A1A9VYH2 A0A0K8V8L3 A0A067RP15 A0A0A1WIV2 A0A1A9Y9J4 W8C398 A0A154PB96 A0A1B0B9E2 A0A1J1J3R8 A0A1A9Z3I8 U4UNY1 A0A1B0FC46 N6ULQ5 A0A151WFB6 A0A195F2S0 F4WM77 A0A195DDQ4 A0A0L7L9J7 A0A034VY99 A0A195BD14 E9IEX7 A0A158P334 A0A151IGD8 A0A1Y1N7A1
PDB
6G53
E-value=0,
Score=1633
Ontologies
GO
PANTHER
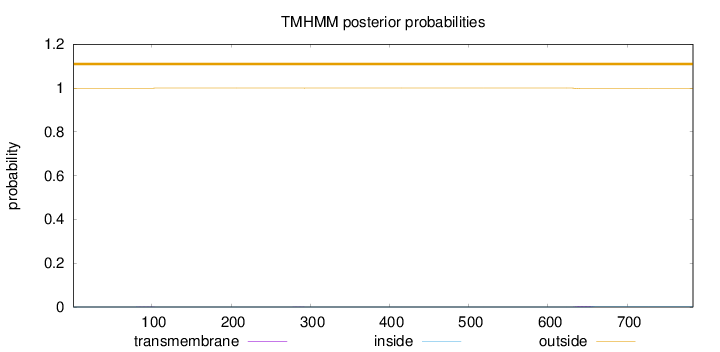
Topology
Subcellular location
Length:
783
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0713999999999999
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00054
outside
1 - 783
Population Genetic Test Statistics
Pi
27.567449
Theta
29.481575
Tajima's D
0.124166
CLR
1.772898
CSRT
0.407029648517574
Interpretation
Uncertain
