Gene
KWMTBOMO12889 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012691
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_catalase-like_[Bombyx_mori]
Full name
Catalase
Location in the cell
Mitochondrial Reliability : 1.213 Peroxisomal Reliability : 1.285
Sequence
CDS
ATGTTGCGTTTGACGATAGCGTTGCTTGCGATGGCCGCGGTGGCAAATACTGGGAACGAGCCAGCGCCGGCAGCCCAGCAGATAGTCGCGTTCAAACAGAGAACATCGGGGCCCATCGGTATTATGACAACGAGTGCTGGATCGCCGATCGAACTCAAAGATGCGAGTAGCACATTGAACTCAAATCTCATATTCAATGAATATTTTATGGATTCATTAACTCATATAGTACGAGAGAGGATTCCTGAGAGGTTGGTGCATGCAAAAGCGGGTGGAGCATTCGGTTATTTTGAGGTTACGCATGATATTACGAATATTTGTAAAGCCAAATTATTTACTAAAATAGGAAAGAAAACCCCAGTGGCAGCAAGATTTTCTCCAGCGGTAGTCGAAAGAGGAGGTACTGACACTTCAAGAGATGCGCGCGGATTTGCTATTAAATTTTACACTGAAGACGGTAATTTTGATATCGTGGGATTTAATACGCCCATGTACTCGTACAAAGATCCACTGCTTTTTTCAGCATTTGTGCGAGCTCAAAAACGAAATCCAGCTACTAATTTATTAGACGGTAATACGCTATGGGACTTTTTGACGTTGCACCCGGAGAGCTTGCATATATTTTTACTCGTATTTGGGGATAGAGGCATACCAGATGGTTACAGACATATGCCAGGGTTTGGCATTCACACTTATCAAGTTGTGAATGAGAGTGGAGAGAAGCACTTTGTTAGATTTCATTTTATTCCTGACGCTGGCATAAAAAATCTAAACTCTAAACGAGCACAGAAAATATCCGGTATCGATCCAGATTACGCCACCAGAGATTTGTATAATGCGATAGGAAAGGGTAAATTTCCGAGCTGGTCTGTCAGTATCCAGATATTGACATTAGACGATGTCAAAAATGCAAAATTCGATGTTTTTGACGTGACAAAAGTATTACCCCAAGATGAATTTCCCCTAAAGCCACTCGGTCGTTTGGTTTTGAACAAAAATCCGACGAACTATTTCGCAGAAATAGAACAGTTGGCGTATAGCCCAGCAAATCTTGTGCCGGGCATTTTGGGAGGACCGGACAAAGTTTTCGAAGCTAGACGCTTAGCGTATAGAGACGCTCAATATTACCGGCTAGGTGCTAACTTTAATAAAATACCGGTAAATTGTCCGATTCAAAATCACGTACTTGCTTACAATCGCGATGGTCGACCTCCCGTTAAGGATAACGGTAAAGATACCCCCAATTATTATCCTAACACGTTCCATGGACCGGTGCCTTACAAAGACGAGAATAGGGTTGACCTTGTCGAGATATACCAAGATGATCCTAACAACTTCGATCAGGCTAGAGAGCTGTATATGAACGAAATGACGAAAGGGGAACGAAGCAGACTAGTCGAAAATATTCTGTATAGTCTTGGCGGTGTTATCGAAGAATTGCAGGATAGGGCAGTGAAACTTTTCACAATCATACACCCAGATCTTGGTTACAGAATATCTCAAGGTCTAAAAGTGAACCGAACGTCGTACTACAGGGATGATGAATAG
Protein
MLRLTIALLAMAAVANTGNEPAPAAQQIVAFKQRTSGPIGIMTTSAGSPIELKDASSTLNSNLIFNEYFMDSLTHIVRERIPERLVHAKAGGAFGYFEVTHDITNICKAKLFTKIGKKTPVAARFSPAVVERGGTDTSRDARGFAIKFYTEDGNFDIVGFNTPMYSYKDPLLFSAFVRAQKRNPATNLLDGNTLWDFLTLHPESLHIFLLVFGDRGIPDGYRHMPGFGIHTYQVVNESGEKHFVRFHFIPDAGIKNLNSKRAQKISGIDPDYATRDLYNAIGKGKFPSWSVSIQILTLDDVKNAKFDVFDVTKVLPQDEFPLKPLGRLVLNKNPTNYFAEIEQLAYSPANLVPGILGGPDKVFEARRLAYRDAQYYRLGANFNKIPVNCPIQNHVLAYNRDGRPPVKDNGKDTPNYYPNTFHGPVPYKDENRVDLVEIYQDDPNNFDQARELYMNEMTKGERSRLVENILYSLGGVIEELQDRAVKLFTIIHPDLGYRISQGLKVNRTSYYRDDE
Summary
Description
Occurs in almost all aerobically respiring organisms and serves to protect cells from the toxic effects of hydrogen peroxide.
Catalytic Activity
2 H2O2 = 2 H2O + O2
Cofactor
heme
Similarity
Belongs to the catalase family.
Uniprot
H9JT29
H9JY90
H9JY91
A0A0N0PDT6
A0A194Q886
A0A0L7L1C7
+ More
A0A2H1V005 A0A2A4JP02 A0A3S2LYY1 A0A212F4X0 A0A0L7L5S1 A0A212F4W1 A0A2A4JNL0 A0A2H1W235 A0A2R2PTF8 A0A1V0QH47 A0A2T7P5M1 A0A0L7KXM9 A0A194PRI4 A0A2H1WQ68 T1FMR4 A0A1L8EEQ5 S4PA78 B4KEW9 A0A1L8EEC3 G5CDD8 H9U5T8 A0A3B0JH35 A0A3B0K782 A0A0M3QTJ1 A0A0M4EF65 A0A2Z4N440 A0A1I8PLT9 J9HGP0 B4M9N1 A0A286RZM9 B3N7D9 A0A212F436 I4DMH7 B4HYQ5 B4Q788 B3MLZ0 A0A0L0C8Y9 A0A2S0RQP9 A0A194QVH0 Q9VLI6 T1E1G6 C0MII7 Q29JU0 B4GJ58 B2XQY0 B3NHN7 B3M869 A0AMY5 H9BEW3 B4NXH2 A0A034WTI4 A0A0C9Q530 A0A2S2PUK2 A0A3R7QHC4 B0WIN9 A0A2S2R3I9 Q16J86 A0A182GFN9 A0A1I8MF15 A0A023EUF0 B4N7T2 A0A1S3JUG5 A0A0K8VAH0 A0A1S4JIQ3 B4JQA3 T1PCG9 A0A2H8TEJ7 H9ITX1 A0A1W4VMC8 A0A0S1SHH8 A0A1I8PLM9 D3TRU3 B4J1T4 A0A0A1WYM5 C0MII2 A0A0N9E6Y9 A0A0M4EK05 Q6R2J1 A0A1L7LQD2 A0A067RLP1 B4IFV9 A0A0L0C232 T2FDK5 A0A1Q3FPN2 K4N1L2 A0A1A9V5X9 Q68AP5 B4QPW5 A0A1B0A740 H8XYP6 A0A1W4UPD3 B4LFX2 J9JLF8
A0A2H1V005 A0A2A4JP02 A0A3S2LYY1 A0A212F4X0 A0A0L7L5S1 A0A212F4W1 A0A2A4JNL0 A0A2H1W235 A0A2R2PTF8 A0A1V0QH47 A0A2T7P5M1 A0A0L7KXM9 A0A194PRI4 A0A2H1WQ68 T1FMR4 A0A1L8EEQ5 S4PA78 B4KEW9 A0A1L8EEC3 G5CDD8 H9U5T8 A0A3B0JH35 A0A3B0K782 A0A0M3QTJ1 A0A0M4EF65 A0A2Z4N440 A0A1I8PLT9 J9HGP0 B4M9N1 A0A286RZM9 B3N7D9 A0A212F436 I4DMH7 B4HYQ5 B4Q788 B3MLZ0 A0A0L0C8Y9 A0A2S0RQP9 A0A194QVH0 Q9VLI6 T1E1G6 C0MII7 Q29JU0 B4GJ58 B2XQY0 B3NHN7 B3M869 A0AMY5 H9BEW3 B4NXH2 A0A034WTI4 A0A0C9Q530 A0A2S2PUK2 A0A3R7QHC4 B0WIN9 A0A2S2R3I9 Q16J86 A0A182GFN9 A0A1I8MF15 A0A023EUF0 B4N7T2 A0A1S3JUG5 A0A0K8VAH0 A0A1S4JIQ3 B4JQA3 T1PCG9 A0A2H8TEJ7 H9ITX1 A0A1W4VMC8 A0A0S1SHH8 A0A1I8PLM9 D3TRU3 B4J1T4 A0A0A1WYM5 C0MII2 A0A0N9E6Y9 A0A0M4EK05 Q6R2J1 A0A1L7LQD2 A0A067RLP1 B4IFV9 A0A0L0C232 T2FDK5 A0A1Q3FPN2 K4N1L2 A0A1A9V5X9 Q68AP5 B4QPW5 A0A1B0A740 H8XYP6 A0A1W4UPD3 B4LFX2 J9JLF8
EC Number
1.11.1.6
Pubmed
19121390
26354079
26227816
22118469
23254933
23622113
+ More
17994087 17510324 22651552 22936249 26108605 10731132 12537568 12537572 12537573 12537574 16110336 16951084 17569856 17569867 19126864 26109357 26109356 24330624 15632085 18353680 17550304 25348373 26483478 25315136 24945155 20353571 25830018 15325332 23142216 26760975 28076409 24845553 15763464 22293093
17994087 17510324 22651552 22936249 26108605 10731132 12537568 12537572 12537573 12537574 16110336 16951084 17569856 17569867 19126864 26109357 26109356 24330624 15632085 18353680 17550304 25348373 26483478 25315136 24945155 20353571 25830018 15325332 23142216 26760975 28076409 24845553 15763464 22293093
EMBL
BABH01030664
BABH01042963
BABH01042965
BABH01042966
KQ460041
KPJ18348.1
+ More
KQ459324 KPJ01604.1 JTDY01003585 KOB69303.1 ODYU01000045 SOQ34149.1 NWSH01000997 PCG73173.1 RSAL01000108 RVE47224.1 AGBW02010296 OWR48787.1 JTDY01002827 KOB70646.1 OWR48788.1 PCG73174.1 ODYU01005857 SOQ47155.1 KX079633 KX065288 ANA84065.1 KY701971 ARE67828.1 PZQS01000006 PVD28719.1 JTDY01004753 KOB67801.1 KQ459594 KPI96061.1 ODYU01010180 SOQ55126.1 AMQM01003159 KB096023 ESO09072.1 GFDG01001766 JAV17033.1 GAIX01003369 JAA89191.1 CH933807 EDW11938.1 GFDG01001805 JAV16994.1 JN051294 AEP40969.1 JQ663444 AFG31725.1 OUUW01000006 SPP81495.1 SPP81496.1 CP012523 ALC39003.1 ALC39005.1 MG397135 AWX63639.1 CH478024 EJY58037.1 CH940654 EDW57907.1 MF737386 ASX95436.1 CH954177 EDV58290.1 AGBW02010446 OWR48497.1 AK402495 BAM19117.1 CH480818 EDW52185.1 CM000361 CM002910 EDX04282.1 KMY89138.1 CH902620 EDV31818.1 JRES01000836 KNC27864.1 MF979105 AWA45968.1 KQ461108 KPJ09319.1 AE014134 AY058324 AM294019 AM294020 AM294021 AM294022 AM294024 AM294025 AM294026 AM294027 FM245150 FM245151 FM245152 FM245153 FM245155 FM245156 FM245157 FM245158 AAF52704.1 AAL13553.1 CAL25923.1 CAL25924.1 CAL25925.1 CAL25926.1 CAL25928.1 CAL25929.1 CAL25930.1 CAL25931.1 CAR93076.1 CAR93077.1 CAR93078.1 CAR93079.1 CAR93081.1 CAR93082.1 CAR93083.1 CAR93084.1 GALA01001689 JAA93163.1 FM245159 CAR93085.1 CH379062 EAL32871.1 CH479184 EDW37372.1 EU102287 ABW82155.1 CH954178 EDV51832.1 CH902618 EDV41008.1 AM294023 CAL25927.1 JQ009332 AFC98367.1 CM000157 EDW88563.1 GAKP01001864 JAC57088.1 GBYB01009173 JAG78940.1 GGMR01020510 MBY33129.1 QCYY01001316 ROT78899.1 DS231951 EDS28616.1 GGMS01015355 MBY84558.1 EAT34333.1 JXUM01010672 JXUM01010673 KQ560314 KXJ83094.1 GAPW01001047 JAC12551.1 CH964214 EDW80421.1 GDHF01016433 JAI35881.1 CH916372 EDV99083.1 KA645638 AFP60267.1 GFXV01000706 MBW12511.1 BABH01007112 KR908786 ALM09355.1 CCAG010022811 EZ424145 ADD20421.1 CH916366 EDV98014.1 GBXI01010123 JAD04169.1 FM245154 CAR93080.1 KR072552 ALF39394.1 CP012525 ALC44426.1 AY518322 JX162772 AAR99908.1 AFM68912.1 FX983163 BAV93783.1 KK852549 KDR21530.1 CH480834 EDW46546.1 JRES01000996 KNC26307.1 KF367466 AGV76059.1 GFDL01005612 JAV29433.1 JX513905 AFV36369.1 AB164195 BAD38853.1 CM000363 CM002912 EDX10993.1 KMZ00447.1 HQ668089 AET34916.1 CH940647 EDW70371.1 ABLF02017906
KQ459324 KPJ01604.1 JTDY01003585 KOB69303.1 ODYU01000045 SOQ34149.1 NWSH01000997 PCG73173.1 RSAL01000108 RVE47224.1 AGBW02010296 OWR48787.1 JTDY01002827 KOB70646.1 OWR48788.1 PCG73174.1 ODYU01005857 SOQ47155.1 KX079633 KX065288 ANA84065.1 KY701971 ARE67828.1 PZQS01000006 PVD28719.1 JTDY01004753 KOB67801.1 KQ459594 KPI96061.1 ODYU01010180 SOQ55126.1 AMQM01003159 KB096023 ESO09072.1 GFDG01001766 JAV17033.1 GAIX01003369 JAA89191.1 CH933807 EDW11938.1 GFDG01001805 JAV16994.1 JN051294 AEP40969.1 JQ663444 AFG31725.1 OUUW01000006 SPP81495.1 SPP81496.1 CP012523 ALC39003.1 ALC39005.1 MG397135 AWX63639.1 CH478024 EJY58037.1 CH940654 EDW57907.1 MF737386 ASX95436.1 CH954177 EDV58290.1 AGBW02010446 OWR48497.1 AK402495 BAM19117.1 CH480818 EDW52185.1 CM000361 CM002910 EDX04282.1 KMY89138.1 CH902620 EDV31818.1 JRES01000836 KNC27864.1 MF979105 AWA45968.1 KQ461108 KPJ09319.1 AE014134 AY058324 AM294019 AM294020 AM294021 AM294022 AM294024 AM294025 AM294026 AM294027 FM245150 FM245151 FM245152 FM245153 FM245155 FM245156 FM245157 FM245158 AAF52704.1 AAL13553.1 CAL25923.1 CAL25924.1 CAL25925.1 CAL25926.1 CAL25928.1 CAL25929.1 CAL25930.1 CAL25931.1 CAR93076.1 CAR93077.1 CAR93078.1 CAR93079.1 CAR93081.1 CAR93082.1 CAR93083.1 CAR93084.1 GALA01001689 JAA93163.1 FM245159 CAR93085.1 CH379062 EAL32871.1 CH479184 EDW37372.1 EU102287 ABW82155.1 CH954178 EDV51832.1 CH902618 EDV41008.1 AM294023 CAL25927.1 JQ009332 AFC98367.1 CM000157 EDW88563.1 GAKP01001864 JAC57088.1 GBYB01009173 JAG78940.1 GGMR01020510 MBY33129.1 QCYY01001316 ROT78899.1 DS231951 EDS28616.1 GGMS01015355 MBY84558.1 EAT34333.1 JXUM01010672 JXUM01010673 KQ560314 KXJ83094.1 GAPW01001047 JAC12551.1 CH964214 EDW80421.1 GDHF01016433 JAI35881.1 CH916372 EDV99083.1 KA645638 AFP60267.1 GFXV01000706 MBW12511.1 BABH01007112 KR908786 ALM09355.1 CCAG010022811 EZ424145 ADD20421.1 CH916366 EDV98014.1 GBXI01010123 JAD04169.1 FM245154 CAR93080.1 KR072552 ALF39394.1 CP012525 ALC44426.1 AY518322 JX162772 AAR99908.1 AFM68912.1 FX983163 BAV93783.1 KK852549 KDR21530.1 CH480834 EDW46546.1 JRES01000996 KNC26307.1 KF367466 AGV76059.1 GFDL01005612 JAV29433.1 JX513905 AFV36369.1 AB164195 BAD38853.1 CM000363 CM002912 EDX10993.1 KMZ00447.1 HQ668089 AET34916.1 CH940647 EDW70371.1 ABLF02017906
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000218220
UP000283053
+ More
UP000007151 UP000245119 UP000015101 UP000009192 UP000268350 UP000092553 UP000095300 UP000008820 UP000008792 UP000008711 UP000001292 UP000000304 UP000007801 UP000037069 UP000000803 UP000001819 UP000008744 UP000002282 UP000283509 UP000002320 UP000069940 UP000249989 UP000095301 UP000007798 UP000085678 UP000001070 UP000192221 UP000092444 UP000027135 UP000078200 UP000092445 UP000007819
UP000007151 UP000245119 UP000015101 UP000009192 UP000268350 UP000092553 UP000095300 UP000008820 UP000008792 UP000008711 UP000001292 UP000000304 UP000007801 UP000037069 UP000000803 UP000001819 UP000008744 UP000002282 UP000283509 UP000002320 UP000069940 UP000249989 UP000095301 UP000007798 UP000085678 UP000001070 UP000192221 UP000092444 UP000027135 UP000078200 UP000092445 UP000007819
Interpro
IPR020835
Catalase_sf
+ More
IPR011614 Catalase_core
IPR024711 Catalase_clade1/3
IPR024708 Catalase_AS
IPR018028 Catalase
IPR010582 Catalase_immune_responsive
IPR037060 Catalase_core_sf
IPR040333 Catalase_3
IPR002226 Catalase_haem_BS
IPR019786 Zinc_finger_PHD-type_CS
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR011011 Znf_FYVE_PHD
IPR015424 PyrdxlP-dep_Trfase
IPR015421 PyrdxlP-dep_Trfase_major
IPR005814 Aminotrans_3
IPR015422 PyrdxlP-dep_Trfase_dom1
IPR011614 Catalase_core
IPR024711 Catalase_clade1/3
IPR024708 Catalase_AS
IPR018028 Catalase
IPR010582 Catalase_immune_responsive
IPR037060 Catalase_core_sf
IPR040333 Catalase_3
IPR002226 Catalase_haem_BS
IPR019786 Zinc_finger_PHD-type_CS
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR011011 Znf_FYVE_PHD
IPR015424 PyrdxlP-dep_Trfase
IPR015421 PyrdxlP-dep_Trfase_major
IPR005814 Aminotrans_3
IPR015422 PyrdxlP-dep_Trfase_dom1
Gene 3D
ProteinModelPortal
H9JT29
H9JY90
H9JY91
A0A0N0PDT6
A0A194Q886
A0A0L7L1C7
+ More
A0A2H1V005 A0A2A4JP02 A0A3S2LYY1 A0A212F4X0 A0A0L7L5S1 A0A212F4W1 A0A2A4JNL0 A0A2H1W235 A0A2R2PTF8 A0A1V0QH47 A0A2T7P5M1 A0A0L7KXM9 A0A194PRI4 A0A2H1WQ68 T1FMR4 A0A1L8EEQ5 S4PA78 B4KEW9 A0A1L8EEC3 G5CDD8 H9U5T8 A0A3B0JH35 A0A3B0K782 A0A0M3QTJ1 A0A0M4EF65 A0A2Z4N440 A0A1I8PLT9 J9HGP0 B4M9N1 A0A286RZM9 B3N7D9 A0A212F436 I4DMH7 B4HYQ5 B4Q788 B3MLZ0 A0A0L0C8Y9 A0A2S0RQP9 A0A194QVH0 Q9VLI6 T1E1G6 C0MII7 Q29JU0 B4GJ58 B2XQY0 B3NHN7 B3M869 A0AMY5 H9BEW3 B4NXH2 A0A034WTI4 A0A0C9Q530 A0A2S2PUK2 A0A3R7QHC4 B0WIN9 A0A2S2R3I9 Q16J86 A0A182GFN9 A0A1I8MF15 A0A023EUF0 B4N7T2 A0A1S3JUG5 A0A0K8VAH0 A0A1S4JIQ3 B4JQA3 T1PCG9 A0A2H8TEJ7 H9ITX1 A0A1W4VMC8 A0A0S1SHH8 A0A1I8PLM9 D3TRU3 B4J1T4 A0A0A1WYM5 C0MII2 A0A0N9E6Y9 A0A0M4EK05 Q6R2J1 A0A1L7LQD2 A0A067RLP1 B4IFV9 A0A0L0C232 T2FDK5 A0A1Q3FPN2 K4N1L2 A0A1A9V5X9 Q68AP5 B4QPW5 A0A1B0A740 H8XYP6 A0A1W4UPD3 B4LFX2 J9JLF8
A0A2H1V005 A0A2A4JP02 A0A3S2LYY1 A0A212F4X0 A0A0L7L5S1 A0A212F4W1 A0A2A4JNL0 A0A2H1W235 A0A2R2PTF8 A0A1V0QH47 A0A2T7P5M1 A0A0L7KXM9 A0A194PRI4 A0A2H1WQ68 T1FMR4 A0A1L8EEQ5 S4PA78 B4KEW9 A0A1L8EEC3 G5CDD8 H9U5T8 A0A3B0JH35 A0A3B0K782 A0A0M3QTJ1 A0A0M4EF65 A0A2Z4N440 A0A1I8PLT9 J9HGP0 B4M9N1 A0A286RZM9 B3N7D9 A0A212F436 I4DMH7 B4HYQ5 B4Q788 B3MLZ0 A0A0L0C8Y9 A0A2S0RQP9 A0A194QVH0 Q9VLI6 T1E1G6 C0MII7 Q29JU0 B4GJ58 B2XQY0 B3NHN7 B3M869 A0AMY5 H9BEW3 B4NXH2 A0A034WTI4 A0A0C9Q530 A0A2S2PUK2 A0A3R7QHC4 B0WIN9 A0A2S2R3I9 Q16J86 A0A182GFN9 A0A1I8MF15 A0A023EUF0 B4N7T2 A0A1S3JUG5 A0A0K8VAH0 A0A1S4JIQ3 B4JQA3 T1PCG9 A0A2H8TEJ7 H9ITX1 A0A1W4VMC8 A0A0S1SHH8 A0A1I8PLM9 D3TRU3 B4J1T4 A0A0A1WYM5 C0MII2 A0A0N9E6Y9 A0A0M4EK05 Q6R2J1 A0A1L7LQD2 A0A067RLP1 B4IFV9 A0A0L0C232 T2FDK5 A0A1Q3FPN2 K4N1L2 A0A1A9V5X9 Q68AP5 B4QPW5 A0A1B0A740 H8XYP6 A0A1W4UPD3 B4LFX2 J9JLF8
PDB
1QQW
E-value=1.37667e-131,
Score=1204
Ontologies
GO
PANTHER
Topology
SignalP
Position: 1 - 15,
Likelihood: 0.967456
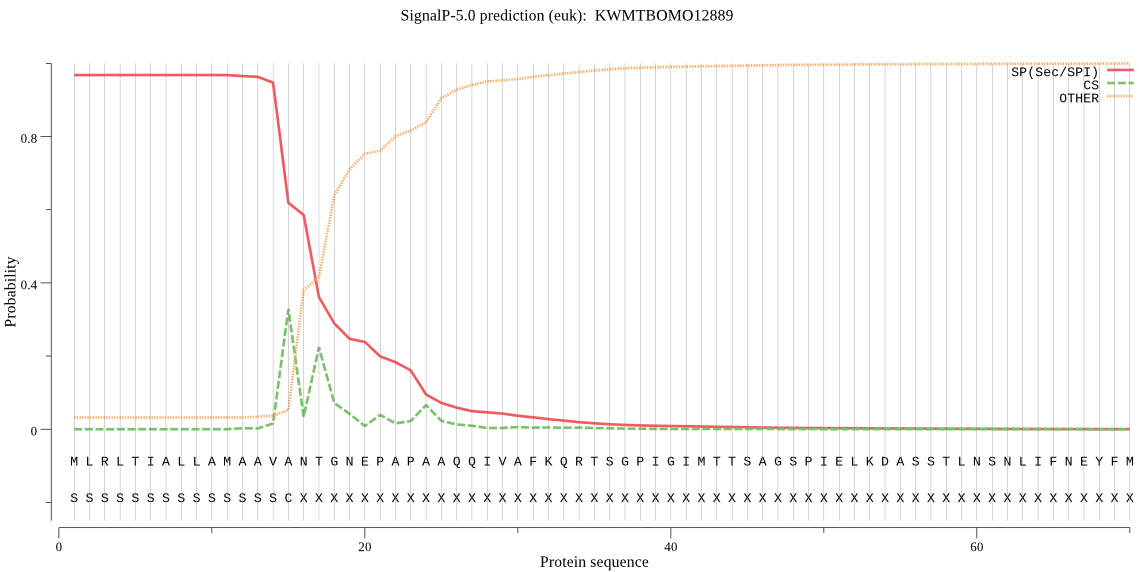
Length:
515
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01614
Exp number, first 60 AAs:
0.01497
Total prob of N-in:
0.00125
outside
1 - 515
Population Genetic Test Statistics
Pi
385.424541
Theta
198.171534
Tajima's D
2.966101
CLR
0.34273
CSRT
0.976951152442378
Interpretation
Uncertain
