Gene
KWMTBOMO12883
Pre Gene Modal
BGIBMGA012694
Annotation
PREDICTED:_phosphatidate_cytidylyltransferase?_photoreceptor-specific_[Amyelois_transitella]
Full name
Phosphatidate cytidylyltransferase
+ More
Phosphatidate cytidylyltransferase, photoreceptor-specific
Phosphatidate cytidylyltransferase, photoreceptor-specific
Alternative Name
CDP-DAG synthase
CDP-DG synthase
CDP-diacylglycerol synthase
CDP-diglyceride pyrophosphorylase
CDP-diglyceride synthase
CTP:phosphatidate cytidylyltransferase
CDP-DG synthase
CDP-diacylglycerol synthase
CDP-diglyceride pyrophosphorylase
CDP-diglyceride synthase
CTP:phosphatidate cytidylyltransferase
Location in the cell
PlasmaMembrane Reliability : 4.971
Sequence
CDS
ATGAGTGAAATAAGGCAACGGCGGGGAGATGGCGATGGAAACCAGAAAATAGAAGCTGCTGTCGAGTCGGATCATGTCGACTCTGAAGAGGAGAAAGTCCTAGAAGAGAAGTATGTGGACGAGCTAGCGAAGTCCTTACCGCAAGGCACTGACAAAACTCCAGAAATCCTTGACTCTGCGCTTTCTGGACTATCTACGAGATGGAGGAACTGGGTGATTCGTGGAATATTTACATGGTTGATGATCGGAGGTTTCTGTTTGCTGATCTACGGTGGACCTCTCGCTCTCATGATTACGGTCCTCTGCGTTCAAGTGAAGTGTTTTGAAGAGATCATCAACATTGGATATGCAGTCTACCGTGTCCATGGTCTCCCATGGTTCCGTTCCTTGTCTTGGTACTTCCTATTGACTTCCAATTATTTCTTCTATGGGGAAAACTTGATTGACTACTTTGGTGTTGTCATCAACAGAACGGACTACCTCAAGTTTCTGGTGACGTACCACCGTTTCATATCGTTCAGCTTGTACTGCGTCGGCTTCGTTTGGTTCGTGCTGTCTCTGGTCAAACGCTACTACATGCGGCAGTTCAGCCTGTTCGCTTGGACCCACGTCGCTCTGCTGATCGTCGTTACGCAGTCGTATCTCATCATCCAGAACATATTTGAGGGTCTGATCTGGTTCATAGTGCCGGTGTCGATGATCATCTGCAATGACGTCATGGCCTACGTATTCGGCTTCTTCTTCGGCAAGACTCCGCTCATCAAACTGAGCCCCAAGAAGACCTGGGAGGGTTTCATTGGCGGCGGTATATCTACAGTCATATTCGGTCTCGGGCTTTCCTATATAATGTCACAATATCCTTACCTTGTTTGTCCCATCGAATACTCCGAGAGTGTGGGCCGCATGATGGACTGCGAGCCCTCGCTGCTGTTCAGACTCCAGGAGTACACGCCGCCTCAATTCCTGCAACCCGTTATGAAAGTGATCGGCATGGACAAGTTCAACATGTACCCCTTCATGATTCACTCGCTGTTCCTGAGCACCTTCAGTTCAGTGATTGGACCGTTCGGTGGTTTCTTCGCTTCCGGATTCAAGAGGGCCTTCAAGATCAAGGACTTCGGAGATGTGATCCCTGGGCACGGCGGCATCATGGACCGCTTCGACTGCCAGTTCCTGATGGCCACCTTCGTCAACGTCTACATCACCAGCTTCATCCGCACCGCTACTCCTCAGAAGCTGCTGCAGCAGGTGTACAACTTGAAGCCGGAACAGCAGCTTCAAATGTTCTACGCCCTTAAGGAGTCCCTGGAACATAGGAATATTCTGAACGCCATACCCTAG
Protein
MSEIRQRRGDGDGNQKIEAAVESDHVDSEEEKVLEEKYVDELAKSLPQGTDKTPEILDSALSGLSTRWRNWVIRGIFTWLMIGGFCLLIYGGPLALMITVLCVQVKCFEEIINIGYAVYRVHGLPWFRSLSWYFLLTSNYFFYGENLIDYFGVVINRTDYLKFLVTYHRFISFSLYCVGFVWFVLSLVKRYYMRQFSLFAWTHVALLIVVTQSYLIIQNIFEGLIWFIVPVSMIICNDVMAYVFGFFFGKTPLIKLSPKKTWEGFIGGGISTVIFGLGLSYIMSQYPYLVCPIEYSESVGRMMDCEPSLLFRLQEYTPPQFLQPVMKVIGMDKFNMYPFMIHSLFLSTFSSVIGPFGGFFASGFKRAFKIKDFGDVIPGHGGIMDRFDCQFLMATFVNVYITSFIRTATPQKLLQQVYNLKPEQQLQMFYALKESLEHRNILNAIP
Summary
Description
Catalyzes the conversion of phosphatidic acid (PA) to CDP-diacylglycerol (CDP-DAG), an important precursor for the synthesis of phosphatidylinositol (PtdIns) and phosphatidylglycerol (PG) (PubMed:7816135, PubMed:24603715). Required for the regeneration of the signaling molecule phosphatidylinositol 4,5-bisphosphate (PtdInsP2) from PA and maintenance of its steady supply during signaling thus playing an essential role during phospholipase C-mediated transduction (PubMed:7816135). In the salivary glands and possibly other adipose tissues, function is essential for regulating cell growth and neutral lipid storage by coordinating PtdIns metabolism and insulin pathway activity (PubMed:24603715). Acts by positively regulating activity of the insulin pathway through synthesis of PtdIns, and in turn the insulin pathway up-regulates the synthesis of CdsA (PubMed:24603715). This CdsA-insulin positive feedback loop may be one of the mechanisms for coordinating cell growth and fat storage; switching to fat storage when cells reach homeostasis or converting from growth to fat storage under nutrient-poor conditions (PubMed:24603715).
Catalytic Activity
a 1,2-diacyl-sn-glycero-3-phosphate + CTP + H(+) = a CDP-1,2-diacyl-sn-glycerol + diphosphate
Similarity
Belongs to the CDS family.
Keywords
Complete proteome
Lipid biosynthesis
Lipid metabolism
Membrane
Nucleotidyltransferase
Phospholipid biosynthesis
Phospholipid metabolism
Phosphoprotein
Reference proteome
Sensory transduction
Transferase
Transmembrane
Transmembrane helix
Vision
Feature
chain Phosphatidate cytidylyltransferase, photoreceptor-specific
Uniprot
A0A3S2NGU7
A0A2A4K0Y5
A0A194Q891
A0A212FDQ1
A0A0N1PHA0
A0A067QR92
+ More
A0A2J7Q7J6 A0A2P8Y1F1 A0A195CGB7 A0A1Q3FKG6 A0A1Q3FKC3 F4X838 A0A195F9H8 A0A158NCB0 A0A151J6A2 A0A1J1J7I7 E2AG77 A0A034W123 A0A0K8U383 A0A1B6IMZ7 V9IGJ0 A0A0C9RIF2 W8B3M9 A0A026WKY4 A0A0L7QP64 A0A1B0CWJ0 A0A1B0GFE8 A0A154PIL1 A0A1A9X0G9 A0A2A3EEX9 K7J483 Q17JP6 A0A1B6LEU8 A0A182WJU2 A0A182PDP4 A0A182RCL3 A0A182Y2N6 A0A232F4I7 A0A182J196 Q1HRB7 A0A182UU30 B0WRX4 A0A182I2M8 A0A182WX36 Q7QJ08 A0A0M8ZNQ7 A0A1D2NM81 A0A1B6G1U6 A0A182FGL9 B4HIZ8 A0A2M4A5V6 B4QLI4 A0A2M4CUT9 B4LFG6 X2JC55 B4PCT2 P56079 H9ZJM9 B3NF51 F3YDP2 A0A2M4BMA5 A0A1B0DAQ0 D6WJ88 A0A2M3ZGH4 A0A1W4VP43 T1P9V2 A0A1B6CB71 A0A023ESH4 V5GV21 A0A182NJX1 Q29FD7 B4H1H0 B4KWY0 B4MXE3 A0A0M5JB16 A0A3B0KFP3 B3M419 A0A0N7ZHF4 E9GA99 A0A1Y1KXL5 U5EIN5 A0A0P5GSH7 A0A1L8DYP2 A0A0A9W898 A0A0J7L254 A0A182R0V8 A0A1W4WHG4 A0A2H8TWY3 A0A151WFL8 B4IWR7 A0A0F7VK29 R4FPG7 A0A224XD54 A0A069DYY0 J9JUZ1 A0A023F925 A0A0V0GC37 N6U2G1 A0A2S2QSM6 B6IDT4
A0A2J7Q7J6 A0A2P8Y1F1 A0A195CGB7 A0A1Q3FKG6 A0A1Q3FKC3 F4X838 A0A195F9H8 A0A158NCB0 A0A151J6A2 A0A1J1J7I7 E2AG77 A0A034W123 A0A0K8U383 A0A1B6IMZ7 V9IGJ0 A0A0C9RIF2 W8B3M9 A0A026WKY4 A0A0L7QP64 A0A1B0CWJ0 A0A1B0GFE8 A0A154PIL1 A0A1A9X0G9 A0A2A3EEX9 K7J483 Q17JP6 A0A1B6LEU8 A0A182WJU2 A0A182PDP4 A0A182RCL3 A0A182Y2N6 A0A232F4I7 A0A182J196 Q1HRB7 A0A182UU30 B0WRX4 A0A182I2M8 A0A182WX36 Q7QJ08 A0A0M8ZNQ7 A0A1D2NM81 A0A1B6G1U6 A0A182FGL9 B4HIZ8 A0A2M4A5V6 B4QLI4 A0A2M4CUT9 B4LFG6 X2JC55 B4PCT2 P56079 H9ZJM9 B3NF51 F3YDP2 A0A2M4BMA5 A0A1B0DAQ0 D6WJ88 A0A2M3ZGH4 A0A1W4VP43 T1P9V2 A0A1B6CB71 A0A023ESH4 V5GV21 A0A182NJX1 Q29FD7 B4H1H0 B4KWY0 B4MXE3 A0A0M5JB16 A0A3B0KFP3 B3M419 A0A0N7ZHF4 E9GA99 A0A1Y1KXL5 U5EIN5 A0A0P5GSH7 A0A1L8DYP2 A0A0A9W898 A0A0J7L254 A0A182R0V8 A0A1W4WHG4 A0A2H8TWY3 A0A151WFL8 B4IWR7 A0A0F7VK29 R4FPG7 A0A224XD54 A0A069DYY0 J9JUZ1 A0A023F925 A0A0V0GC37 N6U2G1 A0A2S2QSM6 B6IDT4
EC Number
2.7.7.41
Pubmed
26354079
22118469
24845553
29403074
21719571
21347285
+ More
20798317 25348373 24495485 24508170 30249741 20075255 17510324 25244985 28648823 17204158 12364791 14747013 17210077 27289101 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 7816135 18327897 24603715 18362917 19820115 25315136 24945155 15632085 18057021 21292972 28004739 25401762 26823975 26334808 25474469 23537049
20798317 25348373 24495485 24508170 30249741 20075255 17510324 25244985 28648823 17204158 12364791 14747013 17210077 27289101 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 7816135 18327897 24603715 18362917 19820115 25315136 24945155 15632085 18057021 21292972 28004739 25401762 26823975 26334808 25474469 23537049
EMBL
RSAL01000108
RVE47228.1
NWSH01000262
PCG77907.1
KQ459324
KPJ01609.1
+ More
AGBW02009028 OWR51828.1 KQ460232 KPJ16413.1 KK853114 KDR11188.1 NEVH01017442 PNF24556.1 PYGN01001050 PSN38085.1 KQ977873 KYM99128.1 GFDL01006966 JAV28079.1 GFDL01006995 JAV28050.1 GL888912 EGI57387.1 KQ981727 KYN37098.1 ADTU01011780 ADTU01011781 KQ979863 KYN18754.1 CVRI01000075 CRL08363.1 GL439266 EFN67568.1 GAKP01010568 JAC48384.1 GDHF01031311 GDHF01014513 JAI21003.1 JAI37801.1 GECU01019423 JAS88283.1 JR046328 AEY60175.1 GBYB01013002 JAG82769.1 GAMC01010823 GAMC01010822 GAMC01010820 JAB95735.1 KK107167 QOIP01000010 EZA56331.1 RLU17805.1 KQ414843 KOC60291.1 AJWK01032487 CCAG010017310 KQ434908 KZC11314.1 KZ288263 PBC30315.1 AAZX01003264 CH477231 EAT46929.1 GEBQ01017749 JAT22228.1 NNAY01001043 OXU25373.1 DQ440177 ABF18210.1 DS232062 EDS33584.1 APCN01000170 AAAB01008807 EAA04188.4 KQ435955 KOX68030.1 LJIJ01000006 ODN06380.1 GECZ01013363 JAS56406.1 CH480815 EDW40652.1 GGFK01002781 MBW36102.1 CM000363 CM002912 EDX09644.1 KMY98285.1 GGFL01004753 MBW68931.1 CH940647 EDW69264.1 AE014296 AHN58001.1 CM000159 EDW93836.1 BT133385 AFH36370.1 CH954178 EDV50393.1 BT126403 AED98583.1 GGFJ01005069 MBW54210.1 AJVK01013365 AJVK01013366 AJVK01013367 KQ971343 EFA04441.1 GGFM01006840 MBW27591.1 KA644728 AFP59357.1 GEDC01026763 GEDC01024211 GEDC01015771 GEDC01011520 GEDC01006408 GEDC01004981 JAS10535.1 JAS13087.1 JAS21527.1 JAS25778.1 JAS30890.1 JAS32317.1 GAPW01001378 JAC12220.1 GALX01000482 JAB67984.1 CH379067 EAL31318.1 CH479202 EDW30147.1 CH933809 EDW18601.1 CH963876 EDW76712.1 CP012525 ALC43431.1 OUUW01000012 SPP87180.1 CH902618 EDV40381.1 GDIP01245105 JAI78296.1 GL732536 EFX83743.1 GEZM01071041 JAV66104.1 GANO01002560 JAB57311.1 GDIQ01259618 JAJ92106.1 GFDF01002672 JAV11412.1 GBHO01040881 GBHO01040880 GBRD01005127 GDHC01011658 GDHC01004225 GDHC01003234 JAG02723.1 JAG02724.1 JAG60694.1 JAQ06971.1 JAQ14404.1 JAQ15395.1 LBMM01001179 KMQ96771.1 AXCN02000532 GFXV01006969 MBW18774.1 KQ983211 KYQ46642.1 CH916366 EDV96293.1 LN830646 CFW94155.1 ACPB03017242 GAHY01000188 JAA77322.1 GFTR01006040 JAW10386.1 GBGD01001605 JAC87284.1 ABLF02036211 GBBI01000741 JAC17971.1 GECL01000444 JAP05680.1 APGK01044857 KB741028 KB632297 ENN74811.1 ERL91694.1 GGMS01011470 MBY80673.1 BT050524 ACJ13231.1
AGBW02009028 OWR51828.1 KQ460232 KPJ16413.1 KK853114 KDR11188.1 NEVH01017442 PNF24556.1 PYGN01001050 PSN38085.1 KQ977873 KYM99128.1 GFDL01006966 JAV28079.1 GFDL01006995 JAV28050.1 GL888912 EGI57387.1 KQ981727 KYN37098.1 ADTU01011780 ADTU01011781 KQ979863 KYN18754.1 CVRI01000075 CRL08363.1 GL439266 EFN67568.1 GAKP01010568 JAC48384.1 GDHF01031311 GDHF01014513 JAI21003.1 JAI37801.1 GECU01019423 JAS88283.1 JR046328 AEY60175.1 GBYB01013002 JAG82769.1 GAMC01010823 GAMC01010822 GAMC01010820 JAB95735.1 KK107167 QOIP01000010 EZA56331.1 RLU17805.1 KQ414843 KOC60291.1 AJWK01032487 CCAG010017310 KQ434908 KZC11314.1 KZ288263 PBC30315.1 AAZX01003264 CH477231 EAT46929.1 GEBQ01017749 JAT22228.1 NNAY01001043 OXU25373.1 DQ440177 ABF18210.1 DS232062 EDS33584.1 APCN01000170 AAAB01008807 EAA04188.4 KQ435955 KOX68030.1 LJIJ01000006 ODN06380.1 GECZ01013363 JAS56406.1 CH480815 EDW40652.1 GGFK01002781 MBW36102.1 CM000363 CM002912 EDX09644.1 KMY98285.1 GGFL01004753 MBW68931.1 CH940647 EDW69264.1 AE014296 AHN58001.1 CM000159 EDW93836.1 BT133385 AFH36370.1 CH954178 EDV50393.1 BT126403 AED98583.1 GGFJ01005069 MBW54210.1 AJVK01013365 AJVK01013366 AJVK01013367 KQ971343 EFA04441.1 GGFM01006840 MBW27591.1 KA644728 AFP59357.1 GEDC01026763 GEDC01024211 GEDC01015771 GEDC01011520 GEDC01006408 GEDC01004981 JAS10535.1 JAS13087.1 JAS21527.1 JAS25778.1 JAS30890.1 JAS32317.1 GAPW01001378 JAC12220.1 GALX01000482 JAB67984.1 CH379067 EAL31318.1 CH479202 EDW30147.1 CH933809 EDW18601.1 CH963876 EDW76712.1 CP012525 ALC43431.1 OUUW01000012 SPP87180.1 CH902618 EDV40381.1 GDIP01245105 JAI78296.1 GL732536 EFX83743.1 GEZM01071041 JAV66104.1 GANO01002560 JAB57311.1 GDIQ01259618 JAJ92106.1 GFDF01002672 JAV11412.1 GBHO01040881 GBHO01040880 GBRD01005127 GDHC01011658 GDHC01004225 GDHC01003234 JAG02723.1 JAG02724.1 JAG60694.1 JAQ06971.1 JAQ14404.1 JAQ15395.1 LBMM01001179 KMQ96771.1 AXCN02000532 GFXV01006969 MBW18774.1 KQ983211 KYQ46642.1 CH916366 EDV96293.1 LN830646 CFW94155.1 ACPB03017242 GAHY01000188 JAA77322.1 GFTR01006040 JAW10386.1 GBGD01001605 JAC87284.1 ABLF02036211 GBBI01000741 JAC17971.1 GECL01000444 JAP05680.1 APGK01044857 KB741028 KB632297 ENN74811.1 ERL91694.1 GGMS01011470 MBY80673.1 BT050524 ACJ13231.1
Proteomes
UP000283053
UP000218220
UP000053268
UP000007151
UP000053240
UP000027135
+ More
UP000235965 UP000245037 UP000078542 UP000007755 UP000078541 UP000005205 UP000078492 UP000183832 UP000000311 UP000053097 UP000279307 UP000053825 UP000092461 UP000092444 UP000076502 UP000091820 UP000242457 UP000002358 UP000008820 UP000075920 UP000075885 UP000075900 UP000076408 UP000215335 UP000075880 UP000075903 UP000002320 UP000075840 UP000076407 UP000007062 UP000053105 UP000094527 UP000069272 UP000001292 UP000000304 UP000008792 UP000000803 UP000002282 UP000008711 UP000092462 UP000007266 UP000192221 UP000095301 UP000075884 UP000001819 UP000008744 UP000009192 UP000007798 UP000092553 UP000268350 UP000007801 UP000000305 UP000036403 UP000075886 UP000192223 UP000075809 UP000001070 UP000015103 UP000007819 UP000019118 UP000030742
UP000235965 UP000245037 UP000078542 UP000007755 UP000078541 UP000005205 UP000078492 UP000183832 UP000000311 UP000053097 UP000279307 UP000053825 UP000092461 UP000092444 UP000076502 UP000091820 UP000242457 UP000002358 UP000008820 UP000075920 UP000075885 UP000075900 UP000076408 UP000215335 UP000075880 UP000075903 UP000002320 UP000075840 UP000076407 UP000007062 UP000053105 UP000094527 UP000069272 UP000001292 UP000000304 UP000008792 UP000000803 UP000002282 UP000008711 UP000092462 UP000007266 UP000192221 UP000095301 UP000075884 UP000001819 UP000008744 UP000009192 UP000007798 UP000092553 UP000268350 UP000007801 UP000000305 UP000036403 UP000075886 UP000192223 UP000075809 UP000001070 UP000015103 UP000007819 UP000019118 UP000030742
Interpro
SUPFAM
SSF54160
SSF54160
Gene 3D
CDD
ProteinModelPortal
A0A3S2NGU7
A0A2A4K0Y5
A0A194Q891
A0A212FDQ1
A0A0N1PHA0
A0A067QR92
+ More
A0A2J7Q7J6 A0A2P8Y1F1 A0A195CGB7 A0A1Q3FKG6 A0A1Q3FKC3 F4X838 A0A195F9H8 A0A158NCB0 A0A151J6A2 A0A1J1J7I7 E2AG77 A0A034W123 A0A0K8U383 A0A1B6IMZ7 V9IGJ0 A0A0C9RIF2 W8B3M9 A0A026WKY4 A0A0L7QP64 A0A1B0CWJ0 A0A1B0GFE8 A0A154PIL1 A0A1A9X0G9 A0A2A3EEX9 K7J483 Q17JP6 A0A1B6LEU8 A0A182WJU2 A0A182PDP4 A0A182RCL3 A0A182Y2N6 A0A232F4I7 A0A182J196 Q1HRB7 A0A182UU30 B0WRX4 A0A182I2M8 A0A182WX36 Q7QJ08 A0A0M8ZNQ7 A0A1D2NM81 A0A1B6G1U6 A0A182FGL9 B4HIZ8 A0A2M4A5V6 B4QLI4 A0A2M4CUT9 B4LFG6 X2JC55 B4PCT2 P56079 H9ZJM9 B3NF51 F3YDP2 A0A2M4BMA5 A0A1B0DAQ0 D6WJ88 A0A2M3ZGH4 A0A1W4VP43 T1P9V2 A0A1B6CB71 A0A023ESH4 V5GV21 A0A182NJX1 Q29FD7 B4H1H0 B4KWY0 B4MXE3 A0A0M5JB16 A0A3B0KFP3 B3M419 A0A0N7ZHF4 E9GA99 A0A1Y1KXL5 U5EIN5 A0A0P5GSH7 A0A1L8DYP2 A0A0A9W898 A0A0J7L254 A0A182R0V8 A0A1W4WHG4 A0A2H8TWY3 A0A151WFL8 B4IWR7 A0A0F7VK29 R4FPG7 A0A224XD54 A0A069DYY0 J9JUZ1 A0A023F925 A0A0V0GC37 N6U2G1 A0A2S2QSM6 B6IDT4
A0A2J7Q7J6 A0A2P8Y1F1 A0A195CGB7 A0A1Q3FKG6 A0A1Q3FKC3 F4X838 A0A195F9H8 A0A158NCB0 A0A151J6A2 A0A1J1J7I7 E2AG77 A0A034W123 A0A0K8U383 A0A1B6IMZ7 V9IGJ0 A0A0C9RIF2 W8B3M9 A0A026WKY4 A0A0L7QP64 A0A1B0CWJ0 A0A1B0GFE8 A0A154PIL1 A0A1A9X0G9 A0A2A3EEX9 K7J483 Q17JP6 A0A1B6LEU8 A0A182WJU2 A0A182PDP4 A0A182RCL3 A0A182Y2N6 A0A232F4I7 A0A182J196 Q1HRB7 A0A182UU30 B0WRX4 A0A182I2M8 A0A182WX36 Q7QJ08 A0A0M8ZNQ7 A0A1D2NM81 A0A1B6G1U6 A0A182FGL9 B4HIZ8 A0A2M4A5V6 B4QLI4 A0A2M4CUT9 B4LFG6 X2JC55 B4PCT2 P56079 H9ZJM9 B3NF51 F3YDP2 A0A2M4BMA5 A0A1B0DAQ0 D6WJ88 A0A2M3ZGH4 A0A1W4VP43 T1P9V2 A0A1B6CB71 A0A023ESH4 V5GV21 A0A182NJX1 Q29FD7 B4H1H0 B4KWY0 B4MXE3 A0A0M5JB16 A0A3B0KFP3 B3M419 A0A0N7ZHF4 E9GA99 A0A1Y1KXL5 U5EIN5 A0A0P5GSH7 A0A1L8DYP2 A0A0A9W898 A0A0J7L254 A0A182R0V8 A0A1W4WHG4 A0A2H8TWY3 A0A151WFL8 B4IWR7 A0A0F7VK29 R4FPG7 A0A224XD54 A0A069DYY0 J9JUZ1 A0A023F925 A0A0V0GC37 N6U2G1 A0A2S2QSM6 B6IDT4
PDB
4Q2G
E-value=2.9973e-05,
Score=114
Ontologies
PATHWAY
GO
PANTHER
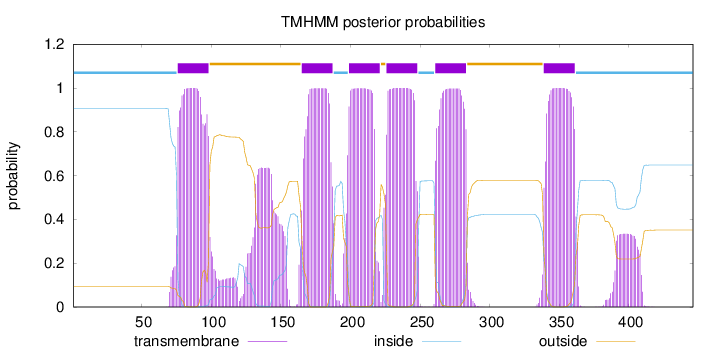
Topology
Subcellular location
Membrane
Length:
446
Number of predicted TMHs:
6
Exp number of AAs in TMHs:
157.07321
Exp number, first 60 AAs:
0
Total prob of N-in:
0.90636
inside
1 - 75
TMhelix
76 - 98
outside
99 - 164
TMhelix
165 - 187
inside
188 - 198
TMhelix
199 - 221
outside
222 - 225
TMhelix
226 - 248
inside
249 - 260
TMhelix
261 - 283
outside
284 - 338
TMhelix
339 - 361
inside
362 - 446
Population Genetic Test Statistics
Pi
194.788407
Theta
170.389893
Tajima's D
0.822148
CLR
0.338377
CSRT
0.609869506524674
Interpretation
Uncertain
