Gene
KWMTBOMO12878 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012701
Annotation
glutamate_synthase_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.67 Nuclear Reliability : 1.012
Sequence
CDS
ATGGAGTGGGAAGGGCCGCCCAAGCAAGGCTTGTACGACCCCCAGAACGAGCACGAAGCCTGCGGTGTTGGTTTTGTTGTTGCTATCGACGGAAAACGCAGTCATAAAATTGTCCGAGATGCTGAAGTCCTGGCTAAACGTATGGAGCACCGCGGTGCCTGCGCGTGCGACAATGATACCGGTGATGGCGCCGGTGTACTGACCGCTATCCCTCATCAATTCTACTGCGCTCAACTAAGAGACAGTCACCAGATCGATTTGCCCCCCTTCGGGCGATATGCGACTGGAATCTTCTTCCTAGACAAACTGCATCACCAGGACATAGAAGCCAAGTTTCTAGTACTAGCCGAGAGTCTTGGTCTTAAGGTGCTGTGCTGGCGTACCGTTCCTACAAAAAACAGCAGCATCGGTCAGGTGGCTCGTAACTCGGAGCCGTATATGCGTCAAGTGTTTGTTACCGGCGATATCGAGGACGAGACCCAGCTGTCGCGGCAGATATTCGTGCTCCGTAAACGCGCAAGCCACGAGCTGGTGGTGCCGGGCGCCAGGTTCTATATATGCAGCCTGTCGCTCAGGACTATCGTGTACAAAGGACTTCTCACTTCTAATCAGCTATGGGAGTACTTCAAGGATCTGAGCAATCCAGCGTTCACAACGTATTTGGCGCTTGTCCACACAAGATTCTCAACGAATACGTTTCCGAGCTGGGAAAGAGCACACCCTTTGAGAGTTCTCGCCCACAATGGAGAAATAAATACGTTACGAGGTAACGTCAATCTGATGAAAGCTCGCGAGGGTGTGATGAAGAGCGACATATTTGGAGACGACTTGAAGAAGCTATATCCAGTGGTGGAGCCCAACCTGTCGGACTCTGGGTCGGCGGACTGCGTGCTGGAGTTCCTGGCGCAGGCGGGCGGCAGGTCGCTGCCCGAGGCCGTGATGACCATGGTGCCCGAGGCGTGGCAGAACGACCACACCATGCACCCCGACAAGCGCGACTACTACCAGTGGGCCTCCTCCGCCATGGAGCCGTGGGACGGACCCGCCCTGGTCTCCTTCACGGACGGCCGGTACATCGGTGCCATACTGGACAGGAACGGTCTGCGGCCGTCTCGTTTCTACGTGACCAGCGAGAACATACTGGTCATGGCCTCCGAAGTCGGAGTGTACGACGTCGAGCCTGAGAAGGTCGTACTCAAGAGTCGTCTGAAGCCGGGACGGATGCTGCTCGTGGACACAGTTCAGAAAAGAATGATTCAAGATGTCGAGTTGAAGATGGAAATAGCGAGGAGCCGACCGCACTCTGAGTGGCTGAGAGATAAGATCACAATGGAGGACATATACAAATCATTGAGTCCAGCCGAGAACGGTTCTACGGTTTCCAACGGCGAATCTCGACCCATTTCGGGCCTTTCTGACAAAAGACTCGGCCTCTTCGGTTACACCATCGAGTCAATCCACATGCTGCTGCTTCCAATGATACAGAACAAGAAGGAAGCCCTCGGAAGCATGGGCAACGACGCTCCGCTGGCGTGCCTGTCCCGCTTCCAGCCGCTGCCCTACGAGTACTTCAAGCAGCTGTTCGCGCAGGTCACGAACCCGCCCATCGATCCGTTCAGGGAGAAGATAGTGATGTCCCTGCTGTGTCCGATCGGACCGGCGCCGAACATCCTGCAGCCCGGCGCGGAGTTCGTGCACCGGCTGTTCCTGCCGCAGCCCGTGCTGTCCCTGCCGGACCTCGACGCGCTCAAGCGGACTAGCTACCGGGGCTGGAAGACGCAAGTATTGGACTGCACGTACGAAGCGTCGTCGGGCGCGAGCGGGCTGGTGGCGGCCCTGAACGCCGTGTGCGCCGCCGCAGAGAGCGCCGCCCGCGCGCGCTGCCAGCTGCTCGTGCTCTCCGACCGCGCCGCCGGACCCACGCGCGTGCCCGTCAGCTCGCTGCTCGCGCTGGGAGCTGTACATCATCATTTAATTGAAACCCGTCAGCGAATGAAAGTTGGACTCATTGTCGAGACGGCCGAAGCGAGAGAGGTTCATCACATGTGCGTTCTGTTAGGCTACGGCGCCGACGCGATCTGTCCGTACTTGGCCTTCGAGCTGGCGTTCGCCCTCCGCAACGACAACATCCTCGACCCGAACCTCACCGACGACGACATCTACTCCGCCTACCAGAAGGCCATTGAGACCGGCCTCGCCAAGGTCATGGCCAAGATGGGCATCAGCACCCTCCAGTCTTACAAGAGCGCTCAGATATTCGAAGCCGTCGGCTTGAGCGAGGAAGTCATCGATAAATGTTTTAAAGGAACCCAATCTAGGATTGGTGGCGTGAACTTTGAGATTCTGTCGAATGAGACGTTGGACCGACACGCCATGACTTACGGGGATTGCGTAGATAGTCTCGTTTTGAGGAACCCGGGCAACTATCACTGGCGCGCCGGCGGAGAGAAGCACGTCAACGACCCCACGTCCATCGCCAACCTGCAGGAGGCCGCGCTCGGGAACAGCAAGTCCGCCTACGACCGGTTCAGAGAGAGCACGCTGGAGTCGGTCCGCGCCTGCACGCTGCGGGGCCAGCTGGAGCTCGTGCGGCTGGACGAGCCCATCCCTATCAGCGAAGTGGAGCCGGCCTCGGAAATCGTTAAACGTTTCGCTACAGGTGCGATGTCATTCGGATCGATATCTCTGGAGGCTCACACGACGCTCGCCATCGCCATGAACCGCATCGGAGGTAAATCCAACACCGGAGAGGGAGGGGAAAACGCTGACCGATATTTGAACCAGGACCCCGACTACAGCAAGCGCTCGGCCATCAAACAAGTAGCGTCGGGACGGTTCGGCGTGACGGCCTCGTACCTCGCGCACGCGGACGATCTGCAGATCAAAATGGCGCAGGGGGCCAAACCCGGGGAAGGTGGAGAACTGCCCGGCTACAAGGTGACGGCGGACATAGCGCGCACGCGGTGCTCGGTGCCGGGCGTGGGGCTGATCTCGCCGCCGCCGCACCACGACATCTACTCCATCGAGGACCTCGCCGAGCTCATCTACGACCTCAAGTGCGCCAACCCCAAGGCTCGCATATCCGTCAAACTGGTGTCCGAGGTCGGGGTCGGGGTCGTGGCTTCCGGGGTCGCCAAGGGCAAGGCGGAGCACATCGTGATCTCGGGGCACGACGGCGGCACGGGCGCGAGCTCGTGGACCGGCATCAAGTCGGCCGGCCTGCCCTGGGAGCTGGGGGTCGCCGAGACGCACCAGGTGCTCGTGCTCAACGACCTCAGATCCAGGGTCGTCGTACAGGCGGACGGTCAGATCCGGACCGGGTTCGACGTGATGGTCGCCGCGCTGCTCGGGGCCGACGAGTTCGGGTTCAGCACCGCGCCGCTCATTGCGCTCGGCTGCACGATGATGCGCAAGTGTCACTTGAACACGTGCCCGGTCGGCATCGCCACGCAGGACCCGGTGCTCCGGAAGAAATTCGCCGGGAAACCTGAACACGTCGTCAACTATGTCTTCATGCTGGTGGAAGAGATACGTCAGCACATGGCCGAGGTCGGTGTGCGGCGCTTCCAAGACCTGATCGGTCGCACGGACCTGCTAAAGGTGCGCGAGAACAACGACAACCCCAAGGCGAGGCTGCTCAACCTGAGCCTCATACTGAAGAACGCTCTCCACATGAGGCCCGGGGTCAACATCATCGGGGGCTCCAAAGCGCAGGACTTCCAACTGGAGAAACGTCTGGACAATCAGCTGATCGAGCAGTGCTCCGGCATCCTGGACGGGACTCAGGCGCACGCCGACATCAAAATGAAAATAACGAACGAGGACCGCGCGTTTACTTCCACCCTGTCCTACCGGATCGCTATGGAATACGGGGACGACGGTCTACCGGACGGTAAAACTGTTAACATCTCATTGACCGGTTCAGCGGGACAAAGCTTCTGCGCGTTCCTGTCCAAGGGCATCACTGTGACGCTCGAGGGAGACGCTAACGACTACGTGGGCAAGGGTCTGTCGGGCGGCACCGTGGTCATCTACCCGCCGCGCGACTCTCCCTTCGAGTCGCATCTCAACGTCATCGTGGGGAACGTGTGTCTGTACGGAGCCACCTCGGGACGCGCCTACTTCAGGGGCATCGCGTCGGAGCGGTTCTGCGTGCGCAACTCTGGCTGCACGGCCGTGGTGGAGGGGGTGGGGGACCACGCGTGCGAGTACATGACGGCGGGCGCGGCGCTCGTGCTGGGGCTCACCGGCAGGAACTTCGCGGCCGGCATGAGCGGTGGCATCGCCTACGTCTACGACATTGATGGTTCATTCAAAGGGAAATGTAATCCTGAAATGGTTGAACTCTTACCTTTGGAACTCGAAGACGACCTCAAATATGTACAGAAACTTCTCGAAGAATTCGTAGAGTACACTGGATCACTGATCGCGGTAGAACTATTAAAGACATGGCCCGAACCAGCGAAGAAGTTCGTCAAAGTGTTTCCGTACGAGTACCAGCGAGCGCTGAAGCAGATGGCTCTGAAGCAGCCGGCCGCGCAGAAGGGCGACACCAATGGACATCAGAACGGCGTGCTCGACATAGAGGAGACCGTTAAGGATGTCGAGTTCGAGAAGAAGAATCTCGAAAAGATCTTGGACAAGACCAGAGGGTTTATGAAGTACGGTCGCGAGACGGCGCTGTACCGCAGCGCGGAGAGCCGGCTGCGCGACTGGGAGGAGATCTACGCGGGGGGTGCGGTGCGGCGCTCCGTGCGCGTGCAGGCGGCGCGCTGCATGGACTGCGGCGTGCCCTTCTGCCAGAGCGCGCACGGCTGCCCGCTCGGCAACCTCATCCCCAAGTGGAACGACCTCATCTACCGCGCCGACTGGGCGCAGGCGCTCGCGCAGCTGCTGCAGACCAACAACTTCCCCGAGTTCACTGGGCGCGTGTGTCCGGCGCCGTGCGAGGGCGCGTGCGTGCTGGCCATCAGCGAGCCCGCCGTCACCATCAAGAACATCGAGTGCGCCATCATCGACCACGCCTTCGAGGCGGGCTGGCTCAAGCCGGAGGTGCCGTCGCATCGCAACGGGCACACGGTCGCCGTGGTGGGCTCGGGCCCCGCGGGGCTGGCCTGCGCGCACCAGCTCAACAAGGCCGGCTACACGGTGACGGTGTTCGAGCGGAACGACCGGGCGGGCGGCCTGCTGCAGTACGGGATACCCACTATGAAGCTGAGCAAGGAGGTCGTCGCCAGAAGGATCAAGCTGATGCAGGCCGAGGGGATCGTCTTCAAGTGCAACGTCGACGTCGGGAAGGATATATCCGCTAATGAATTGGCAAGCGAGTACGACGCGCTGGTGCTGTGCCTGGGCGCGACGTGGCCCCGCGACCTGCCCCTGAAGGGCCGCCAGCTCAACGGCATCCACTACGCCATGGAGTTCCTCGAGACCTGGCAGAAGAAGCAGGCGGGATCCACGCATCCGCACATGAAGGACAACCCCGAACTCAACGCCAAAGATAAGGACGTGCTCATCATCGGAGGCGGAGATACCGGATGCGATTGCATAGCCACTTCGTTGCGTCAGGGGGCGAAGACAATAACGACATTCGAAATCTTACCCGAGCCGAAAAAGACCAGAGGACTAGATAACCCTTGGCCTCAGTGGCCTAGAGTTTTCAGAGTGGACTACGGTCACGAAGAGGTGAAGGTCAAGTTCGGTAACGACCCCAGGAGGTTTTCGACGCTCACCAAAGAATTTTTGGACGACGGCGAGGGCAACGTGTGCGGCGTGCTGGCGGTGGAGGTGGAGTGGAGCCGCGGCGCGGGCGGGCGCTGGGACATGCGCGAGGTGGCGGGCTCGCAGCGGGAGTACCGCGCCGACCTCGTGCTGCTCGCCATGGGCTTCCTCGGCCCCGAGAGATACGTCGCCAACCAGCTCGATCTCCCGCTGGATGCGCGCAGTAACATCGAAACACTGAAATCGGATATTTACAAAACGAACGCGAAGAACGTTTTCGCGGCTGGAGATTGCCGGAGAGGCCAGTCGCTGGTGGTGTGGGCGATATCCGAAGGTCGACAAGCGGCGCGGTCCGTGGACATCTTCCTGTCGGGCCGGTCGTCCCTGCCCGGACCGGGCGGGGTCATACACGAGAACTGA
Protein
MEWEGPPKQGLYDPQNEHEACGVGFVVAIDGKRSHKIVRDAEVLAKRMEHRGACACDNDTGDGAGVLTAIPHQFYCAQLRDSHQIDLPPFGRYATGIFFLDKLHHQDIEAKFLVLAESLGLKVLCWRTVPTKNSSIGQVARNSEPYMRQVFVTGDIEDETQLSRQIFVLRKRASHELVVPGARFYICSLSLRTIVYKGLLTSNQLWEYFKDLSNPAFTTYLALVHTRFSTNTFPSWERAHPLRVLAHNGEINTLRGNVNLMKAREGVMKSDIFGDDLKKLYPVVEPNLSDSGSADCVLEFLAQAGGRSLPEAVMTMVPEAWQNDHTMHPDKRDYYQWASSAMEPWDGPALVSFTDGRYIGAILDRNGLRPSRFYVTSENILVMASEVGVYDVEPEKVVLKSRLKPGRMLLVDTVQKRMIQDVELKMEIARSRPHSEWLRDKITMEDIYKSLSPAENGSTVSNGESRPISGLSDKRLGLFGYTIESIHMLLLPMIQNKKEALGSMGNDAPLACLSRFQPLPYEYFKQLFAQVTNPPIDPFREKIVMSLLCPIGPAPNILQPGAEFVHRLFLPQPVLSLPDLDALKRTSYRGWKTQVLDCTYEASSGASGLVAALNAVCAAAESAARARCQLLVLSDRAAGPTRVPVSSLLALGAVHHHLIETRQRMKVGLIVETAEAREVHHMCVLLGYGADAICPYLAFELAFALRNDNILDPNLTDDDIYSAYQKAIETGLAKVMAKMGISTLQSYKSAQIFEAVGLSEEVIDKCFKGTQSRIGGVNFEILSNETLDRHAMTYGDCVDSLVLRNPGNYHWRAGGEKHVNDPTSIANLQEAALGNSKSAYDRFRESTLESVRACTLRGQLELVRLDEPIPISEVEPASEIVKRFATGAMSFGSISLEAHTTLAIAMNRIGGKSNTGEGGENADRYLNQDPDYSKRSAIKQVASGRFGVTASYLAHADDLQIKMAQGAKPGEGGELPGYKVTADIARTRCSVPGVGLISPPPHHDIYSIEDLAELIYDLKCANPKARISVKLVSEVGVGVVASGVAKGKAEHIVISGHDGGTGASSWTGIKSAGLPWELGVAETHQVLVLNDLRSRVVVQADGQIRTGFDVMVAALLGADEFGFSTAPLIALGCTMMRKCHLNTCPVGIATQDPVLRKKFAGKPEHVVNYVFMLVEEIRQHMAEVGVRRFQDLIGRTDLLKVRENNDNPKARLLNLSLILKNALHMRPGVNIIGGSKAQDFQLEKRLDNQLIEQCSGILDGTQAHADIKMKITNEDRAFTSTLSYRIAMEYGDDGLPDGKTVNISLTGSAGQSFCAFLSKGITVTLEGDANDYVGKGLSGGTVVIYPPRDSPFESHLNVIVGNVCLYGATSGRAYFRGIASERFCVRNSGCTAVVEGVGDHACEYMTAGAALVLGLTGRNFAAGMSGGIAYVYDIDGSFKGKCNPEMVELLPLELEDDLKYVQKLLEEFVEYTGSLIAVELLKTWPEPAKKFVKVFPYEYQRALKQMALKQPAAQKGDTNGHQNGVLDIEETVKDVEFEKKNLEKILDKTRGFMKYGRETALYRSAESRLRDWEEIYAGGAVRRSVRVQAARCMDCGVPFCQSAHGCPLGNLIPKWNDLIYRADWAQALAQLLQTNNFPEFTGRVCPAPCEGACVLAISEPAVTIKNIECAIIDHAFEAGWLKPEVPSHRNGHTVAVVGSGPAGLACAHQLNKAGYTVTVFERNDRAGGLLQYGIPTMKLSKEVVARRIKLMQAEGIVFKCNVDVGKDISANELASEYDALVLCLGATWPRDLPLKGRQLNGIHYAMEFLETWQKKQAGSTHPHMKDNPELNAKDKDVLIIGGGDTGCDCIATSLRQGAKTITTFEILPEPKKTRGLDNPWPQWPRVFRVDYGHEEVKVKFGNDPRRFSTLTKEFLDDGEGNVCGVLAVEVEWSRGAGGRWDMREVAGSQREYRADLVLLAMGFLGPERYVANQLDLPLDARSNIETLKSDIYKTNAKNVFAAGDCRRGQSLVVWAISEGRQAARSVDIFLSGRSSLPGPGGVIHEN
Summary
Cofactor
[3Fe-4S] cluster
Uniprot
Q0KIX8
H9JT39
A0A3S2LYR8
A0A194Q896
A0A212FDN4
A0A2H1V889
+ More
A0A2A4IUS7 A0A2J7QSE7 A0A067QZK1 D6WK43 V5GI16 A0A1S6KZN6 A0A182MWR1 A0A0P4VLV2 A0A182HI12 A0A453YSM0 A0A182URF2 A0A195DNP0 F4W679 A0A195AW55 A0A3L8DBH3 A0A158P206 A0A182RI89 A0A146KRB7 U4U326 E9IS98 A0A182YR31 N6TRN8 A0A182GH21 K7IRG1 A0A411G7W1 A0A023EWE8 A0A182GEZ4 A0A195FJ98 Q16FG2 A0A1Q3FGP6 Q2KQ96 A0A232F2M1 Q7Q5M1 B0WKB3 A0A1Q3FGS2 A0A182FH82 A0A1J1IC53 A0A084WEJ6 A0A2M3YXH0 T1I8Q5 A0A2A3E321 A0A1Q3FGV0 V9IBK9 J9JJH1 W5JK50 A0A1I8P9A7 A0A1D2NM12 A0A182JM70 A0A1W4W5R5 B3M4D8 A0A1W4WHH0 A0A0K8TT44 S5NZT3 B4KY01 A0A0P8ZVG0 B4PK97 E2BBQ1 A0A1W4UTT2 A0A0Q9XDF5 B4LG59 A0A0J9RWP5 B3NDH4 A0A0R1DYB0 Q9VVA4 A0A0M4EH60 A0A0J9RWZ4 A0A1W4UGV3 A0A0Q5UJV8 A0A0Q9WTY0 A0A1L8E2N8 M9NFH8 A0A1L8E2E5 A0A1I8MRP9 T1PDG1 B4N3A9 A0A1I8MRR7 A0A224X4T7 B4IXG2 A0A1B6CX73 A0A1B0G6I1 A0A2U9K363 A0A1A9V4F1 A0A3B0KGL1 A0A1B0AQZ3 A0A1A9YG15 Q29EX1 A0A1B6DVI3 A0A0R3P8Z8 E0VR49 A0A1A9WQ11 A0A034VEZ1 A0A0K8UYB0 A0A226E241 A0A182WTR6
A0A2A4IUS7 A0A2J7QSE7 A0A067QZK1 D6WK43 V5GI16 A0A1S6KZN6 A0A182MWR1 A0A0P4VLV2 A0A182HI12 A0A453YSM0 A0A182URF2 A0A195DNP0 F4W679 A0A195AW55 A0A3L8DBH3 A0A158P206 A0A182RI89 A0A146KRB7 U4U326 E9IS98 A0A182YR31 N6TRN8 A0A182GH21 K7IRG1 A0A411G7W1 A0A023EWE8 A0A182GEZ4 A0A195FJ98 Q16FG2 A0A1Q3FGP6 Q2KQ96 A0A232F2M1 Q7Q5M1 B0WKB3 A0A1Q3FGS2 A0A182FH82 A0A1J1IC53 A0A084WEJ6 A0A2M3YXH0 T1I8Q5 A0A2A3E321 A0A1Q3FGV0 V9IBK9 J9JJH1 W5JK50 A0A1I8P9A7 A0A1D2NM12 A0A182JM70 A0A1W4W5R5 B3M4D8 A0A1W4WHH0 A0A0K8TT44 S5NZT3 B4KY01 A0A0P8ZVG0 B4PK97 E2BBQ1 A0A1W4UTT2 A0A0Q9XDF5 B4LG59 A0A0J9RWP5 B3NDH4 A0A0R1DYB0 Q9VVA4 A0A0M4EH60 A0A0J9RWZ4 A0A1W4UGV3 A0A0Q5UJV8 A0A0Q9WTY0 A0A1L8E2N8 M9NFH8 A0A1L8E2E5 A0A1I8MRP9 T1PDG1 B4N3A9 A0A1I8MRR7 A0A224X4T7 B4IXG2 A0A1B6CX73 A0A1B0G6I1 A0A2U9K363 A0A1A9V4F1 A0A3B0KGL1 A0A1B0AQZ3 A0A1A9YG15 Q29EX1 A0A1B6DVI3 A0A0R3P8Z8 E0VR49 A0A1A9WQ11 A0A034VEZ1 A0A0K8UYB0 A0A226E241 A0A182WTR6
Pubmed
19121390
26354079
22118469
24845553
18362917
19820115
+ More
24330026 27129103 12364791 21719571 30249741 21347285 26823975 23537049 21282665 25244985 26483478 20075255 24945155 17510324 15804581 16876704 28648823 24438588 20920257 23761445 27289101 17994087 26369729 23791183 18057021 17550304 20798317 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 15632085 20566863 25348373
24330026 27129103 12364791 21719571 30249741 21347285 26823975 23537049 21282665 25244985 26483478 20075255 24945155 17510324 15804581 16876704 28648823 24438588 20920257 23761445 27289101 17994087 26369729 23791183 18057021 17550304 20798317 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 15632085 20566863 25348373
EMBL
AB271689
BAF30425.1
BABH01030712
BABH01030713
RSAL01000108
RVE47232.1
+ More
KQ459324 KPJ01614.1 AGBW02009028 OWR51823.1 ODYU01001186 SOQ37055.1 NWSH01007632 PCG62883.1 NEVH01011876 PNF31497.1 KK853069 KDR11809.1 KQ971343 EFA03092.1 GALX01004777 JAB63689.1 KC223373 AQT27182.1 AXCM01001406 AXCM01001407 GDKW01001136 JAI55459.1 APCN01005860 AAAB01008960 KQ980724 KYN14109.1 GL887707 EGI70214.1 KQ976731 KYM76287.1 QOIP01000010 RLU17847.1 ADTU01006925 GDHC01019575 JAP99053.1 KB631854 ERL86743.1 GL765324 EFZ16548.1 APGK01054218 APGK01054219 KB741248 ENN71930.1 JXUM01011025 JXUM01011026 JXUM01011027 JXUM01011028 KQ560322 KXJ83057.1 MH366174 QBB01983.1 GAPW01000003 JAC13595.1 JXUM01010641 JXUM01010642 JXUM01010643 KQ981523 KYN40441.1 CH478447 EAT32975.1 GFDL01008319 JAV26726.1 DQ383822 AAV31916.2 NNAY01001234 OXU24667.1 EAA10819.4 DS231969 EDS29704.1 GFDL01008310 JAV26735.1 CVRI01000047 CRK97779.1 ATLV01023219 KE525341 KFB48640.1 GGFM01000208 MBW20959.1 ACPB03000006 KZ288445 PBC25656.1 GFDL01008292 JAV26753.1 JR037586 JR037587 AEY57856.1 AEY57857.1 ABLF02036900 ADMH02000991 ETN64496.1 LJIJ01000007 ODN06304.1 CH902618 EDV40432.1 GDAI01000076 JAI17527.1 KF021986 AGR65733.1 CH933809 EDW18703.1 KPU78570.1 KPU78571.1 KPU78573.1 CM000159 EDW94795.1 KRK02062.1 GL447151 EFN86827.1 KRG06323.1 KRG06325.1 CH940647 EDW69367.1 CM002912 KMZ00111.1 KMZ00114.1 CH954178 EDV52036.1 KQS44083.1 KRK02063.1 KRK02064.1 AE014296 AY094734 AAF49409.2 AAM11087.1 CP012525 ALC43001.1 KMZ00112.1 KMZ00113.1 KMZ00115.1 KQS44082.1 KQS44084.1 KRF84340.1 KRF84343.1 GFDF01001195 JAV12889.1 AFH04475.1 AGB94666.1 GFDF01001196 JAV12888.1 KA646796 KA646797 AFP61425.1 CH964062 EDW78848.2 GFTR01008920 JAW07506.1 CH916366 EDV96399.1 GEDC01019277 JAS18021.1 CCAG010008841 MF446850 AWS20468.1 OUUW01000012 SPP87550.1 JXJN01002171 JXJN01002172 CH379070 EAL29938.1 GEDC01007621 JAS29677.1 KRT08502.1 DS235451 EEB15855.1 GAKP01018613 GAKP01018612 JAC40340.1 GDHF01020665 GDHF01007415 JAI31649.1 JAI44899.1 LNIX01000009 OXA50546.1
KQ459324 KPJ01614.1 AGBW02009028 OWR51823.1 ODYU01001186 SOQ37055.1 NWSH01007632 PCG62883.1 NEVH01011876 PNF31497.1 KK853069 KDR11809.1 KQ971343 EFA03092.1 GALX01004777 JAB63689.1 KC223373 AQT27182.1 AXCM01001406 AXCM01001407 GDKW01001136 JAI55459.1 APCN01005860 AAAB01008960 KQ980724 KYN14109.1 GL887707 EGI70214.1 KQ976731 KYM76287.1 QOIP01000010 RLU17847.1 ADTU01006925 GDHC01019575 JAP99053.1 KB631854 ERL86743.1 GL765324 EFZ16548.1 APGK01054218 APGK01054219 KB741248 ENN71930.1 JXUM01011025 JXUM01011026 JXUM01011027 JXUM01011028 KQ560322 KXJ83057.1 MH366174 QBB01983.1 GAPW01000003 JAC13595.1 JXUM01010641 JXUM01010642 JXUM01010643 KQ981523 KYN40441.1 CH478447 EAT32975.1 GFDL01008319 JAV26726.1 DQ383822 AAV31916.2 NNAY01001234 OXU24667.1 EAA10819.4 DS231969 EDS29704.1 GFDL01008310 JAV26735.1 CVRI01000047 CRK97779.1 ATLV01023219 KE525341 KFB48640.1 GGFM01000208 MBW20959.1 ACPB03000006 KZ288445 PBC25656.1 GFDL01008292 JAV26753.1 JR037586 JR037587 AEY57856.1 AEY57857.1 ABLF02036900 ADMH02000991 ETN64496.1 LJIJ01000007 ODN06304.1 CH902618 EDV40432.1 GDAI01000076 JAI17527.1 KF021986 AGR65733.1 CH933809 EDW18703.1 KPU78570.1 KPU78571.1 KPU78573.1 CM000159 EDW94795.1 KRK02062.1 GL447151 EFN86827.1 KRG06323.1 KRG06325.1 CH940647 EDW69367.1 CM002912 KMZ00111.1 KMZ00114.1 CH954178 EDV52036.1 KQS44083.1 KRK02063.1 KRK02064.1 AE014296 AY094734 AAF49409.2 AAM11087.1 CP012525 ALC43001.1 KMZ00112.1 KMZ00113.1 KMZ00115.1 KQS44082.1 KQS44084.1 KRF84340.1 KRF84343.1 GFDF01001195 JAV12889.1 AFH04475.1 AGB94666.1 GFDF01001196 JAV12888.1 KA646796 KA646797 AFP61425.1 CH964062 EDW78848.2 GFTR01008920 JAW07506.1 CH916366 EDV96399.1 GEDC01019277 JAS18021.1 CCAG010008841 MF446850 AWS20468.1 OUUW01000012 SPP87550.1 JXJN01002171 JXJN01002172 CH379070 EAL29938.1 GEDC01007621 JAS29677.1 KRT08502.1 DS235451 EEB15855.1 GAKP01018613 GAKP01018612 JAC40340.1 GDHF01020665 GDHF01007415 JAI31649.1 JAI44899.1 LNIX01000009 OXA50546.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000007151
UP000218220
UP000235965
+ More
UP000027135 UP000007266 UP000075883 UP000075840 UP000075903 UP000078492 UP000007755 UP000078540 UP000279307 UP000005205 UP000075900 UP000030742 UP000076408 UP000019118 UP000069940 UP000249989 UP000002358 UP000078541 UP000008820 UP000215335 UP000007062 UP000002320 UP000069272 UP000183832 UP000030765 UP000015103 UP000242457 UP000007819 UP000000673 UP000095300 UP000094527 UP000075880 UP000192223 UP000007801 UP000009192 UP000002282 UP000008237 UP000192221 UP000008792 UP000008711 UP000000803 UP000092553 UP000095301 UP000007798 UP000001070 UP000092444 UP000078200 UP000268350 UP000092460 UP000092443 UP000001819 UP000009046 UP000091820 UP000198287 UP000076407
UP000027135 UP000007266 UP000075883 UP000075840 UP000075903 UP000078492 UP000007755 UP000078540 UP000279307 UP000005205 UP000075900 UP000030742 UP000076408 UP000019118 UP000069940 UP000249989 UP000002358 UP000078541 UP000008820 UP000215335 UP000007062 UP000002320 UP000069272 UP000183832 UP000030765 UP000015103 UP000242457 UP000007819 UP000000673 UP000095300 UP000094527 UP000075880 UP000192223 UP000007801 UP000009192 UP000002282 UP000008237 UP000192221 UP000008792 UP000008711 UP000000803 UP000092553 UP000095301 UP000007798 UP000001070 UP000092444 UP000078200 UP000268350 UP000092460 UP000092443 UP000001819 UP000009046 UP000091820 UP000198287 UP000076407
Pfam
Interpro
IPR009051
Helical_ferredxn
+ More
IPR006005 Glut_synth_ssu1
IPR006982 Glu_synth_centr_N
IPR012220 Glu_synth_euk
IPR002932 Glu_synthdom
IPR036188 FAD/NAD-bd_sf
IPR028261 DPD_II
IPR036485 Glu_synth_asu_C_sf
IPR013785 Aldolase_TIM
IPR017932 GATase_2_dom
IPR029055 Ntn_hydrolases_N
IPR023753 FAD/NAD-binding_dom
IPR002489 Glu_synth_asu_C
IPR006005 Glut_synth_ssu1
IPR006982 Glu_synth_centr_N
IPR012220 Glu_synth_euk
IPR002932 Glu_synthdom
IPR036188 FAD/NAD-bd_sf
IPR028261 DPD_II
IPR036485 Glu_synth_asu_C_sf
IPR013785 Aldolase_TIM
IPR017932 GATase_2_dom
IPR029055 Ntn_hydrolases_N
IPR023753 FAD/NAD-binding_dom
IPR002489 Glu_synth_asu_C
ProteinModelPortal
Q0KIX8
H9JT39
A0A3S2LYR8
A0A194Q896
A0A212FDN4
A0A2H1V889
+ More
A0A2A4IUS7 A0A2J7QSE7 A0A067QZK1 D6WK43 V5GI16 A0A1S6KZN6 A0A182MWR1 A0A0P4VLV2 A0A182HI12 A0A453YSM0 A0A182URF2 A0A195DNP0 F4W679 A0A195AW55 A0A3L8DBH3 A0A158P206 A0A182RI89 A0A146KRB7 U4U326 E9IS98 A0A182YR31 N6TRN8 A0A182GH21 K7IRG1 A0A411G7W1 A0A023EWE8 A0A182GEZ4 A0A195FJ98 Q16FG2 A0A1Q3FGP6 Q2KQ96 A0A232F2M1 Q7Q5M1 B0WKB3 A0A1Q3FGS2 A0A182FH82 A0A1J1IC53 A0A084WEJ6 A0A2M3YXH0 T1I8Q5 A0A2A3E321 A0A1Q3FGV0 V9IBK9 J9JJH1 W5JK50 A0A1I8P9A7 A0A1D2NM12 A0A182JM70 A0A1W4W5R5 B3M4D8 A0A1W4WHH0 A0A0K8TT44 S5NZT3 B4KY01 A0A0P8ZVG0 B4PK97 E2BBQ1 A0A1W4UTT2 A0A0Q9XDF5 B4LG59 A0A0J9RWP5 B3NDH4 A0A0R1DYB0 Q9VVA4 A0A0M4EH60 A0A0J9RWZ4 A0A1W4UGV3 A0A0Q5UJV8 A0A0Q9WTY0 A0A1L8E2N8 M9NFH8 A0A1L8E2E5 A0A1I8MRP9 T1PDG1 B4N3A9 A0A1I8MRR7 A0A224X4T7 B4IXG2 A0A1B6CX73 A0A1B0G6I1 A0A2U9K363 A0A1A9V4F1 A0A3B0KGL1 A0A1B0AQZ3 A0A1A9YG15 Q29EX1 A0A1B6DVI3 A0A0R3P8Z8 E0VR49 A0A1A9WQ11 A0A034VEZ1 A0A0K8UYB0 A0A226E241 A0A182WTR6
A0A2A4IUS7 A0A2J7QSE7 A0A067QZK1 D6WK43 V5GI16 A0A1S6KZN6 A0A182MWR1 A0A0P4VLV2 A0A182HI12 A0A453YSM0 A0A182URF2 A0A195DNP0 F4W679 A0A195AW55 A0A3L8DBH3 A0A158P206 A0A182RI89 A0A146KRB7 U4U326 E9IS98 A0A182YR31 N6TRN8 A0A182GH21 K7IRG1 A0A411G7W1 A0A023EWE8 A0A182GEZ4 A0A195FJ98 Q16FG2 A0A1Q3FGP6 Q2KQ96 A0A232F2M1 Q7Q5M1 B0WKB3 A0A1Q3FGS2 A0A182FH82 A0A1J1IC53 A0A084WEJ6 A0A2M3YXH0 T1I8Q5 A0A2A3E321 A0A1Q3FGV0 V9IBK9 J9JJH1 W5JK50 A0A1I8P9A7 A0A1D2NM12 A0A182JM70 A0A1W4W5R5 B3M4D8 A0A1W4WHH0 A0A0K8TT44 S5NZT3 B4KY01 A0A0P8ZVG0 B4PK97 E2BBQ1 A0A1W4UTT2 A0A0Q9XDF5 B4LG59 A0A0J9RWP5 B3NDH4 A0A0R1DYB0 Q9VVA4 A0A0M4EH60 A0A0J9RWZ4 A0A1W4UGV3 A0A0Q5UJV8 A0A0Q9WTY0 A0A1L8E2N8 M9NFH8 A0A1L8E2E5 A0A1I8MRP9 T1PDG1 B4N3A9 A0A1I8MRR7 A0A224X4T7 B4IXG2 A0A1B6CX73 A0A1B0G6I1 A0A2U9K363 A0A1A9V4F1 A0A3B0KGL1 A0A1B0AQZ3 A0A1A9YG15 Q29EX1 A0A1B6DVI3 A0A0R3P8Z8 E0VR49 A0A1A9WQ11 A0A034VEZ1 A0A0K8UYB0 A0A226E241 A0A182WTR6
PDB
1OFE
E-value=0,
Score=2840
Ontologies
PATHWAY
GO
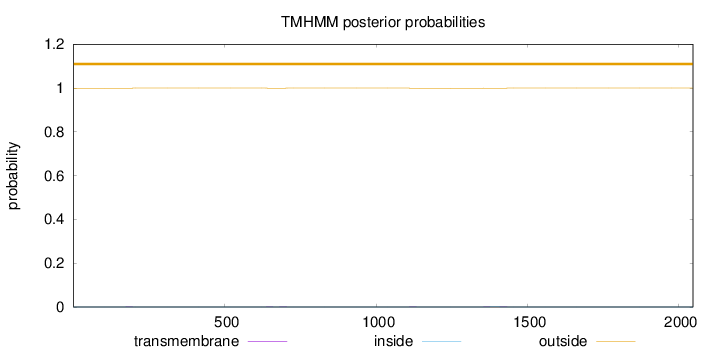
Topology
Length:
2046
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0687700000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00048
outside
1 - 2046
Population Genetic Test Statistics
Pi
204.923594
Theta
171.094246
Tajima's D
0.354685
CLR
0.490023
CSRT
0.472026398680066
Interpretation
Possibly Positive selection
