Gene
KWMTBOMO12868 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012674
Annotation
mRNA_cap-binding_protein_eIF4E_[Bombyx_mori]
Full name
Eukaryotic translation initiation factor 4E
Alternative Name
mRNA cap-binding protein
Location in the cell
Cytoplasmic Reliability : 3.838
Sequence
CDS
ATGGCTGGAAATACTACAGAAGAAGTAGAGGGGTCTACTACCACTGAAACGAAGACCAACAGTAACGCGGAGGTGCCCCCTGAATTTTTGATCAAGCACCCGTTACAAAACCAGTGGAGTCTCTGGTTTTACGACAATGACCGAAACAAAACATGGGAGGAGAATCTTATTGAGTTGACTACATTCGACACAGTTGAAGACTTTTGGAGATTGTACCATCATATTAAGCTACCGTCAGAACTTCGCCAAGGTCATGACTATGCAGTATTCAAGCAAGGCATTCGTCCTATGTGGGAAGACGATGCTAACAAGATGGGAGGAAGATGGCTTATCAGTCTTGAGAAAAAACAACGCTTTACTGATTTAGATAGGTTCTGGTTAGATGTGGTTCTTCTTTTAATTGGTGAAAATTTCGAGAATTCAGATGAGATTTGTGGTGCTGTTGTTAATGTCAGACCAAAAGTAGATAAAATTGCTATTTGGACAGCTGATGCAATGAAGCAGCATGCGACTATTGAAATTGGGAAAAAACTGAAAGAACAGCTAGGGATACACGGCAAGATTGGTTTCCAAGTACACAGAGACACAATGGTTAAGCATAGTTCTGCGACCAAGAATCTATACACTGTTTAG
Protein
MAGNTTEEVEGSTTTETKTNSNAEVPPEFLIKHPLQNQWSLWFYDNDRNKTWEENLIELTTFDTVEDFWRLYHHIKLPSELRQGHDYAVFKQGIRPMWEDDANKMGGRWLISLEKKQRFTDLDRFWLDVVLLLIGENFENSDEICGAVVNVRPKVDKIAIWTADAMKQHATIEIGKKLKEQLGIHGKIGFQVHRDTMVKHSSATKNLYTV
Summary
Description
Recognizes and binds the 7-methylguanosine-containing mRNA cap during an early step in the initiation of protein synthesis and facilitates ribosome binding by inducing the unwinding of the mRNAs secondary structures. Component of the CYFIP1-EIF4E-FMR1 complex which binds to the mRNA cap and mediates translational repression. In the CYFIP1-EIF4E-FMR1 complex this subunit mediates the binding to the mRNA cap (By similarity).
Subunit
eIF4F is a multi-subunit complex, the composition of which varies with external and internal environmental conditions. It is composed of at least EIF4A, EIF4E and EIF4G1/EIF4G3. EIF4E is also known to interact with other partners. The interaction with EIF4ENIF1 mediates the import into the nucleus. Hypophosphorylated EIF4EBP1, EIF4EBP2 and EIF4EBP3 compete with EIF4G1/EIF4G3 to interact with EIF4E; insulin stimulated MAP-kinase (MAPK1 and MAPK3) phosphorylation of EIF4EBP1 causes dissociation of the complex allowing EIF4G1/EIF4G3 to bind and consequent initiation of translation (By similarity). Rapamycin can attenuate insulin stimulation, mediated by FKBPs. Interacts mutually exclusive with EIF4A1 or EIF4A2. Interacts with NGDN and PIWIL2. Component of the CYFIP1-EIF4E-FMR1 complex composed of CYFIP, EIF4E and FMR1. Interacts directly with CYFIP1. Interacts with CLOCK (By similarity). Binds to MKNK2 in nucleus. Interacts with LIMD1, WTIP and AJUBA. Interacts with APOBEC3G in an RNA-dependent manner. Interacts with LARP1. Interacts with METTL3 (By similarity). Interacts with RBM24; this interaction prevents EIF4E from binding to p53/TP53 mRNA and inhibits the assembly of translation initiation complex (By similarity).
Similarity
Belongs to the eukaryotic initiation factor 4E family.
Keywords
Acetylation
Complete proteome
Cytoplasm
Initiation factor
Phosphoprotein
Protein biosynthesis
Reference proteome
RNA-binding
Translation regulation
Feature
chain Eukaryotic translation initiation factor 4E
Uniprot
Q1HPI1
H9JT12
A0A3S2L776
Q964Q9
A0A2A4J1X6
A0A194RBB2
+ More
A0A194Q7W7 I4DIV4 A0A212EVY6 A0A0L7KXB8 F4X4J7 A0A195BDW1 A0A158NIK5 A0A151XFX1 A0A151JUN6 A0A151J2T1 A0A067QP74 E1ZYG8 E2BKQ9 A0A154NZK4 D6WJL4 A0A232F4T7 A0A2J7Q7I4 A0A0J7N7S4 A0A411G752 A0A2P8Y1J2 A0A2A3EFQ9 V9IJ38 A0A088AW55 A0A026W9Z0 A0A1Y1K7E4 A0A3L8DDP4 N6TAG4 A0A0M9A7G6 A0A151IJ30 T1IXQ5 J3JWY7 U5EUM7 A0A0M3TCE3 A0A1B6HUS2 A0A1B6DE09 A0A0A9X0Y9 A0A0K8TSA0 V4A1E3 A0A1B6ERR2 Q17JP2 Q17JP1 A0A0P4WMQ3 A0A0E9X8B5 A0A2R7VS18 A0A2Y9D418 B5DG04 A0A3B4DAX8 W5MSN2 B9EL09 A0A060VY39 A0A0P7WSQ5 D7REL4 A0A2U9CGI3 A9JT06 C1FY44 A0A1Z5KWR6 V9KN06 A0A2U9CCQ9 A0A0Q9WZF9 B8A6A1 A0A2R5LIK9 A0A444UN53 A0A3B3SAF5 B4MX95 A0A401PQS6 A0A3Q1FKS6 A0A1L8EIU2 A0A1L8EIG2 A0A1J1J7F9 A0A3B3WGS5 A0A087XAH6 A0A3B3VFU4 A0A3P9Q183 Q9N0T5 A0A2I4CRN8 A0A023EJP0 A0A3Q3KGT5 U6DU90 W8BCK9 A0A2P6KH27 A0A1A8GGZ9 A0A091ID53 A0A3P9Q1S7 A0A3P8YCK8 A0A0P8XVM0 A0A1L8EB67 C3Y029 A0A1A8FFF2 G3NYE6 A0A2R9CSB0 A0A3B3SAC9 A0A3B0JXP4 A0A3P8V793 I3M004
A0A194Q7W7 I4DIV4 A0A212EVY6 A0A0L7KXB8 F4X4J7 A0A195BDW1 A0A158NIK5 A0A151XFX1 A0A151JUN6 A0A151J2T1 A0A067QP74 E1ZYG8 E2BKQ9 A0A154NZK4 D6WJL4 A0A232F4T7 A0A2J7Q7I4 A0A0J7N7S4 A0A411G752 A0A2P8Y1J2 A0A2A3EFQ9 V9IJ38 A0A088AW55 A0A026W9Z0 A0A1Y1K7E4 A0A3L8DDP4 N6TAG4 A0A0M9A7G6 A0A151IJ30 T1IXQ5 J3JWY7 U5EUM7 A0A0M3TCE3 A0A1B6HUS2 A0A1B6DE09 A0A0A9X0Y9 A0A0K8TSA0 V4A1E3 A0A1B6ERR2 Q17JP2 Q17JP1 A0A0P4WMQ3 A0A0E9X8B5 A0A2R7VS18 A0A2Y9D418 B5DG04 A0A3B4DAX8 W5MSN2 B9EL09 A0A060VY39 A0A0P7WSQ5 D7REL4 A0A2U9CGI3 A9JT06 C1FY44 A0A1Z5KWR6 V9KN06 A0A2U9CCQ9 A0A0Q9WZF9 B8A6A1 A0A2R5LIK9 A0A444UN53 A0A3B3SAF5 B4MX95 A0A401PQS6 A0A3Q1FKS6 A0A1L8EIU2 A0A1L8EIG2 A0A1J1J7F9 A0A3B3WGS5 A0A087XAH6 A0A3B3VFU4 A0A3P9Q183 Q9N0T5 A0A2I4CRN8 A0A023EJP0 A0A3Q3KGT5 U6DU90 W8BCK9 A0A2P6KH27 A0A1A8GGZ9 A0A091ID53 A0A3P9Q1S7 A0A3P8YCK8 A0A0P8XVM0 A0A1L8EB67 C3Y029 A0A1A8FFF2 G3NYE6 A0A2R9CSB0 A0A3B3SAC9 A0A3B0JXP4 A0A3P8V793 I3M004
Pubmed
19121390
11437917
26354079
22651552
22118469
26227816
+ More
21719571 21347285 24845553 20798317 18362917 19820115 28648823 29403074 24508170 28004739 30249741 23537049 22516182 25401762 26823975 26369729 23254933 17510324 25613341 20566863 19878547 20433749 24755649 16191198 23594743 28528879 24402279 17994087 29240929 30297745 11913777 24945155 24495485 25069045 18563158 22722832 24487278
21719571 21347285 24845553 20798317 18362917 19820115 28648823 29403074 24508170 28004739 30249741 23537049 22516182 25401762 26823975 26369729 23254933 17510324 25613341 20566863 19878547 20433749 24755649 16191198 23594743 28528879 24402279 17994087 29240929 30297745 11913777 24945155 24495485 25069045 18563158 22722832 24487278
EMBL
DQ443192
DQ443421
ABF51281.1
ABF51510.1
BABH01030746
RSAL01000108
+ More
RVE47241.1 AF281654 ODYU01010467 AAK94897.1 SOQ55600.1 NWSH01003925 PCG65678.1 KQ460416 KPJ14907.1 KQ459324 KPJ01632.1 AK401222 BAM17844.1 AGBW02012108 OWR45631.1 JTDY01004841 KOB67686.1 GL888656 EGI58627.1 KQ976513 KYM82392.1 ADTU01016896 KQ982179 KYQ59284.1 KQ981733 KYN36548.1 KQ980336 KYN16486.1 KK853114 KDR11192.1 GL435215 EFN73765.1 GL448819 EFN83757.1 KQ434786 KZC05033.1 KQ971342 EFA04625.1 NNAY01000966 OXU25695.1 NEVH01017442 PNF24542.1 LBMM01008684 KMQ88700.1 MH365907 QBB01716.1 PYGN01001050 PSN38116.1 KZ288263 PBC30324.1 JR049929 AEY61128.1 KK107320 EZA52788.1 GEZM01093769 JAV56140.1 QOIP01000009 RLU18441.1 APGK01037209 KB740948 KB632348 ENN77264.1 ERL93182.1 KQ435727 KOX78026.1 KQ977412 KYN02907.1 JH431661 BT127755 AEE62717.1 GANO01003807 JAB56064.1 KR075864 ALE20583.1 GECU01029315 JAS78391.1 GEDC01020118 GEDC01013412 JAS17180.1 JAS23886.1 GBHO01029990 GBRD01011665 GDHC01011361 JAG13614.1 JAG54159.1 JAQ07268.1 GDAI01000580 JAI17023.1 KB202953 ESO87111.1 GECZ01029169 JAS40600.1 CH477231 EAT46933.1 EAT46932.1 GDRN01007367 JAI67949.1 GBXM01009743 JAH98834.1 KK854024 PTY09641.1 AAZO01007334 BT043563 ACH70678.1 AHAT01003979 BT056334 ACM08206.1 FR904332 CDQ59742.1 JARO02005466 KPP66741.1 HM051158 ADH59743.1 CP026257 AWP13982.1 BX927098 BC155151 AAI55152.1 DP001097 ACO88999.1 GFJQ02007562 JAV99407.1 JW867026 AFO99543.1 AWP13983.1 CH963876 KRF98552.1 GGLE01005236 MBY09362.1 SCEB01214206 RXM36605.1 EDW76928.1 BFAA01013763 GCB75471.1 GFDG01000286 JAV18513.1 GFDG01000287 JAV18512.1 CVRI01000075 CRL08365.1 AYCK01011211 AF257235 BC111272 GAPW01004346 GAPW01004345 JAC09252.1 HAAF01014713 CCP86535.1 GAMC01011802 JAB94753.1 MWRG01012060 PRD25635.1 HAEC01002226 SBQ70303.1 KL218441 KFP05295.1 CH902618 KPU78789.1 GFDG01002940 JAV15859.1 GG666477 EEN66395.1 HAEB01011603 SBQ58130.1 AJFE02104908 AJFE02104909 AJFE02104910 AJFE02104911 AJFE02104912 AJFE02104913 OUUW01000002 SPP77491.1 AGTP01041617
RVE47241.1 AF281654 ODYU01010467 AAK94897.1 SOQ55600.1 NWSH01003925 PCG65678.1 KQ460416 KPJ14907.1 KQ459324 KPJ01632.1 AK401222 BAM17844.1 AGBW02012108 OWR45631.1 JTDY01004841 KOB67686.1 GL888656 EGI58627.1 KQ976513 KYM82392.1 ADTU01016896 KQ982179 KYQ59284.1 KQ981733 KYN36548.1 KQ980336 KYN16486.1 KK853114 KDR11192.1 GL435215 EFN73765.1 GL448819 EFN83757.1 KQ434786 KZC05033.1 KQ971342 EFA04625.1 NNAY01000966 OXU25695.1 NEVH01017442 PNF24542.1 LBMM01008684 KMQ88700.1 MH365907 QBB01716.1 PYGN01001050 PSN38116.1 KZ288263 PBC30324.1 JR049929 AEY61128.1 KK107320 EZA52788.1 GEZM01093769 JAV56140.1 QOIP01000009 RLU18441.1 APGK01037209 KB740948 KB632348 ENN77264.1 ERL93182.1 KQ435727 KOX78026.1 KQ977412 KYN02907.1 JH431661 BT127755 AEE62717.1 GANO01003807 JAB56064.1 KR075864 ALE20583.1 GECU01029315 JAS78391.1 GEDC01020118 GEDC01013412 JAS17180.1 JAS23886.1 GBHO01029990 GBRD01011665 GDHC01011361 JAG13614.1 JAG54159.1 JAQ07268.1 GDAI01000580 JAI17023.1 KB202953 ESO87111.1 GECZ01029169 JAS40600.1 CH477231 EAT46933.1 EAT46932.1 GDRN01007367 JAI67949.1 GBXM01009743 JAH98834.1 KK854024 PTY09641.1 AAZO01007334 BT043563 ACH70678.1 AHAT01003979 BT056334 ACM08206.1 FR904332 CDQ59742.1 JARO02005466 KPP66741.1 HM051158 ADH59743.1 CP026257 AWP13982.1 BX927098 BC155151 AAI55152.1 DP001097 ACO88999.1 GFJQ02007562 JAV99407.1 JW867026 AFO99543.1 AWP13983.1 CH963876 KRF98552.1 GGLE01005236 MBY09362.1 SCEB01214206 RXM36605.1 EDW76928.1 BFAA01013763 GCB75471.1 GFDG01000286 JAV18513.1 GFDG01000287 JAV18512.1 CVRI01000075 CRL08365.1 AYCK01011211 AF257235 BC111272 GAPW01004346 GAPW01004345 JAC09252.1 HAAF01014713 CCP86535.1 GAMC01011802 JAB94753.1 MWRG01012060 PRD25635.1 HAEC01002226 SBQ70303.1 KL218441 KFP05295.1 CH902618 KPU78789.1 GFDG01002940 JAV15859.1 GG666477 EEN66395.1 HAEB01011603 SBQ58130.1 AJFE02104908 AJFE02104909 AJFE02104910 AJFE02104911 AJFE02104912 AJFE02104913 OUUW01000002 SPP77491.1 AGTP01041617
Proteomes
UP000005204
UP000283053
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000007755 UP000078540 UP000005205 UP000075809 UP000078541 UP000078492 UP000027135 UP000000311 UP000008237 UP000076502 UP000007266 UP000215335 UP000235965 UP000036403 UP000245037 UP000242457 UP000005203 UP000053097 UP000279307 UP000019118 UP000030742 UP000053105 UP000078542 UP000030746 UP000008820 UP000087266 UP000261440 UP000018468 UP000193380 UP000034805 UP000192224 UP000246464 UP000000437 UP000007798 UP000261540 UP000288216 UP000257200 UP000183832 UP000261480 UP000028760 UP000261500 UP000242638 UP000009136 UP000192220 UP000261640 UP000054308 UP000265140 UP000007801 UP000001554 UP000007635 UP000240080 UP000268350 UP000265120 UP000005215
UP000037510 UP000007755 UP000078540 UP000005205 UP000075809 UP000078541 UP000078492 UP000027135 UP000000311 UP000008237 UP000076502 UP000007266 UP000215335 UP000235965 UP000036403 UP000245037 UP000242457 UP000005203 UP000053097 UP000279307 UP000019118 UP000030742 UP000053105 UP000078542 UP000030746 UP000008820 UP000087266 UP000261440 UP000018468 UP000193380 UP000034805 UP000192224 UP000246464 UP000000437 UP000007798 UP000261540 UP000288216 UP000257200 UP000183832 UP000261480 UP000028760 UP000261500 UP000242638 UP000009136 UP000192220 UP000261640 UP000054308 UP000265140 UP000007801 UP000001554 UP000007635 UP000240080 UP000268350 UP000265120 UP000005215
Interpro
Gene 3D
ProteinModelPortal
Q1HPI1
H9JT12
A0A3S2L776
Q964Q9
A0A2A4J1X6
A0A194RBB2
+ More
A0A194Q7W7 I4DIV4 A0A212EVY6 A0A0L7KXB8 F4X4J7 A0A195BDW1 A0A158NIK5 A0A151XFX1 A0A151JUN6 A0A151J2T1 A0A067QP74 E1ZYG8 E2BKQ9 A0A154NZK4 D6WJL4 A0A232F4T7 A0A2J7Q7I4 A0A0J7N7S4 A0A411G752 A0A2P8Y1J2 A0A2A3EFQ9 V9IJ38 A0A088AW55 A0A026W9Z0 A0A1Y1K7E4 A0A3L8DDP4 N6TAG4 A0A0M9A7G6 A0A151IJ30 T1IXQ5 J3JWY7 U5EUM7 A0A0M3TCE3 A0A1B6HUS2 A0A1B6DE09 A0A0A9X0Y9 A0A0K8TSA0 V4A1E3 A0A1B6ERR2 Q17JP2 Q17JP1 A0A0P4WMQ3 A0A0E9X8B5 A0A2R7VS18 A0A2Y9D418 B5DG04 A0A3B4DAX8 W5MSN2 B9EL09 A0A060VY39 A0A0P7WSQ5 D7REL4 A0A2U9CGI3 A9JT06 C1FY44 A0A1Z5KWR6 V9KN06 A0A2U9CCQ9 A0A0Q9WZF9 B8A6A1 A0A2R5LIK9 A0A444UN53 A0A3B3SAF5 B4MX95 A0A401PQS6 A0A3Q1FKS6 A0A1L8EIU2 A0A1L8EIG2 A0A1J1J7F9 A0A3B3WGS5 A0A087XAH6 A0A3B3VFU4 A0A3P9Q183 Q9N0T5 A0A2I4CRN8 A0A023EJP0 A0A3Q3KGT5 U6DU90 W8BCK9 A0A2P6KH27 A0A1A8GGZ9 A0A091ID53 A0A3P9Q1S7 A0A3P8YCK8 A0A0P8XVM0 A0A1L8EB67 C3Y029 A0A1A8FFF2 G3NYE6 A0A2R9CSB0 A0A3B3SAC9 A0A3B0JXP4 A0A3P8V793 I3M004
A0A194Q7W7 I4DIV4 A0A212EVY6 A0A0L7KXB8 F4X4J7 A0A195BDW1 A0A158NIK5 A0A151XFX1 A0A151JUN6 A0A151J2T1 A0A067QP74 E1ZYG8 E2BKQ9 A0A154NZK4 D6WJL4 A0A232F4T7 A0A2J7Q7I4 A0A0J7N7S4 A0A411G752 A0A2P8Y1J2 A0A2A3EFQ9 V9IJ38 A0A088AW55 A0A026W9Z0 A0A1Y1K7E4 A0A3L8DDP4 N6TAG4 A0A0M9A7G6 A0A151IJ30 T1IXQ5 J3JWY7 U5EUM7 A0A0M3TCE3 A0A1B6HUS2 A0A1B6DE09 A0A0A9X0Y9 A0A0K8TSA0 V4A1E3 A0A1B6ERR2 Q17JP2 Q17JP1 A0A0P4WMQ3 A0A0E9X8B5 A0A2R7VS18 A0A2Y9D418 B5DG04 A0A3B4DAX8 W5MSN2 B9EL09 A0A060VY39 A0A0P7WSQ5 D7REL4 A0A2U9CGI3 A9JT06 C1FY44 A0A1Z5KWR6 V9KN06 A0A2U9CCQ9 A0A0Q9WZF9 B8A6A1 A0A2R5LIK9 A0A444UN53 A0A3B3SAF5 B4MX95 A0A401PQS6 A0A3Q1FKS6 A0A1L8EIU2 A0A1L8EIG2 A0A1J1J7F9 A0A3B3WGS5 A0A087XAH6 A0A3B3VFU4 A0A3P9Q183 Q9N0T5 A0A2I4CRN8 A0A023EJP0 A0A3Q3KGT5 U6DU90 W8BCK9 A0A2P6KH27 A0A1A8GGZ9 A0A091ID53 A0A3P9Q1S7 A0A3P8YCK8 A0A0P8XVM0 A0A1L8EB67 C3Y029 A0A1A8FFF2 G3NYE6 A0A2R9CSB0 A0A3B3SAC9 A0A3B0JXP4 A0A3P8V793 I3M004
PDB
4DUM
E-value=1.55114e-59,
Score=578
Ontologies
PATHWAY
GO
GO:0005737
GO:0003743
GO:0000340
GO:0016281
GO:0046983
GO:0048471
GO:0031370
GO:0019899
GO:0070491
GO:0006417
GO:0005845
GO:0000932
GO:0010494
GO:0005829
GO:0000082
GO:0001662
GO:0019827
GO:0033391
GO:0016442
GO:0045665
GO:0098978
GO:0045931
GO:0071549
GO:0098794
GO:0099578
GO:0017148
GO:0003723
GO:0006413
GO:0015074
GO:0030429
GO:0004003
GO:0005634
GO:0006289
GO:0016286
PANTHER
Topology
Subcellular location
Cytoplasm
P-body
P-body
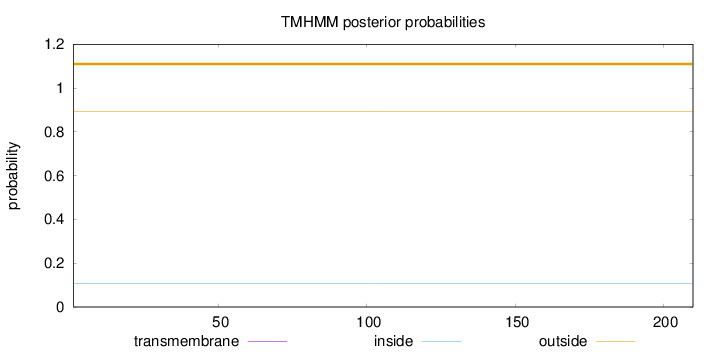
Length:
210
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00023
Exp number, first 60 AAs:
0
Total prob of N-in:
0.10640
outside
1 - 210
Population Genetic Test Statistics
Pi
227.791599
Theta
207.526761
Tajima's D
0.471467
CLR
0.391793
CSRT
0.500474976251187
Interpretation
Uncertain
