Gene
KWMTBOMO12865
Pre Gene Modal
BGIBMGA012709
Annotation
akirin_protein_[Bombyx_mori]
Full name
Akirin
Alternative Name
Protein bhringi
Location in the cell
Nuclear Reliability : 4.234
Sequence
CDS
ATGGCGTGTGCTACATTAAAAAGAAATCTGGATTGGGAGTCTAAGGCGCAATTACCTACAAAGAGAAGAAGATGTTCACCATTTGCAGCAAGTCCAAGCACAAGTCCTGGGTTAAAAACATCAGAATCGAAACCATCTTCATTTGGAGAATCCGTTAGTGCACCTGTGAAAATTACCCCAGAACGCATGGCACAAGAGATTTATGATGAGATTAAACGACTGCATAGACGTGGACAGCTGCGCCTGGCCAACGGCTCTGCTGCATCATGCTCATCATCAAGTGGATCCGAAGGAGACTGTTCACCGCCTCATCAATCAGCTCATGGCCCACAACGTGCCCGCACTCGTGCACTATTCACTTTTAAACAGGTTCGTATGATCTGTGAGCGAATGCTCCATGATCAGGAGGTGGCTCTACGTGCTGAGTATGAGTCTGTACTCAGCACCAAGCTTGCTGAACAGTATGAAGCCTTTGTGAGGTTCAACCTGGATCAGGTGCAGCGCCGACCCCCGCCCAGCACGTGCATGTCGCTCGGTATGGACGCCGAACACATGCACCAGGACCTCGTACCCAGCTATCTGTCCTAA
Protein
MACATLKRNLDWESKAQLPTKRRRCSPFAASPSTSPGLKTSESKPSSFGESVSAPVKITPERMAQEIYDEIKRLHRRGQLRLANGSAASCSSSSGSEGDCSPPHQSAHGPQRARTRALFTFKQVRMICERMLHDQEVALRAEYESVLSTKLAEQYEAFVRFNLDQVQRRPPPSTCMSLGMDAEHMHQDLVPSYLS
Summary
Description
Required for embryonic development and for normal innate immune response. Effector of immune deficiency pathway (Imd) acting either downstream of, or at the level of, the NF-kappa-B factor Relish (Rel). Not part of the Toll pathway (PubMed:18066067). NF-kappa-B factor Relish (Rel) cofactor that orchestrates NF-kappa-B factor Relish (Rel) transcriptional selectivity by recruiting the Osa-containing-SWI/SNF-like Brahma complex (BAP) through bap60 interaction, leading to activation a subset of NF-kappa-B factor Relish (Rel) effector genes (PubMed:25180232).
Subunit
Interacts with dmap1 (PubMed:24947515). Interacts with bap60 and rel; interaction is immune stimulation-dependent; activates selected rel target gene promoters (PubMed:25180232). Interacts with bap55; interaction is immune stimulation-dependent (PubMed:25180232).
Miscellaneous
'Akiraka ni suru' means 'making things clear' in Japanese. The name is given based on the presence of the clear nuclear localization signal.
Similarity
Belongs to the akirin family.
Keywords
Complete proteome
Developmental protein
Immunity
Innate immunity
Nucleus
Phosphoprotein
Reference proteome
Feature
chain Akirin
Uniprot
H6WS69
H9JT47
S4NXV2
A0A2A4IWW2
A0A212EVV2
A0A194RCA2
+ More
A0A194Q9G3 A0A0K8TNC4 U5EWM8 B0X0M8 A0A1J1IB17 T1DF49 T1E2E5 A0A1L8DCY5 T1E2G9 A0A182PIV6 A0A182WI31 A0A023EJ88 A0A1L3N235 A0A182Y884 A0A1L3N228 A0A182R6Q1 A0A182VLD0 A0A182L3Z9 A0A182X279 Q7QIE8 A0A182I6D9 A0A182H6N2 C3RX28 A0A182QLK0 A0A1L3N225 A0A182N7F5 A0A2M4CTL6 A0A2M4AHW6 A0A182FFJ8 A0A1I8MWN9 A0A2M3Z691 T1PBY3 A0A1Q3F078 A0A182J5N6 A0A1I8NMV6 A0A0L0CF21 B4KYP4 A0A1B0DFL2 B4HV59 B4QKD7 M9MRW4 Q9VS59 B4PK68 B3NFE2 Q2M142 B4ML70 B4LDW6 A0A0M4E8R9 Q16MV0 S4UAH5 A0A1L3N221 A0A0A1WHV1 A0A1A9WCW0 B3M5R2 A0A3B0J618 A0A1W4UNU3 A0A1A9VMU2 A0A1A9Z8W3 D3TSF1 A0A0M9A5Y4 W8BNC5 A0A034WPL1 A0A0C9QIA2 A0A0K8VQS4 A0A2A3EBB0 V9IJI1 A0A088AHT4 E5L9J4 A0A1A9Y9Y9 A0A1B0BQ76 A0A154PTI5 K7IUT5 A0A1B6L1T5 A0A1B6GEC8 A0A1B6I263 A0A0L7RIM6 A0A1B6JQA6 A0A067R299 A0A0C9QZ52 D6WLM5 A0A2P8XZD6 A0A0T6B914 A0A1B6DP15 B4J2H1 N6UJ91 A0A232FBL3 A0A1Y1JWR6 A0A1E1X1X1 A0A0L8IF12
A0A194Q9G3 A0A0K8TNC4 U5EWM8 B0X0M8 A0A1J1IB17 T1DF49 T1E2E5 A0A1L8DCY5 T1E2G9 A0A182PIV6 A0A182WI31 A0A023EJ88 A0A1L3N235 A0A182Y884 A0A1L3N228 A0A182R6Q1 A0A182VLD0 A0A182L3Z9 A0A182X279 Q7QIE8 A0A182I6D9 A0A182H6N2 C3RX28 A0A182QLK0 A0A1L3N225 A0A182N7F5 A0A2M4CTL6 A0A2M4AHW6 A0A182FFJ8 A0A1I8MWN9 A0A2M3Z691 T1PBY3 A0A1Q3F078 A0A182J5N6 A0A1I8NMV6 A0A0L0CF21 B4KYP4 A0A1B0DFL2 B4HV59 B4QKD7 M9MRW4 Q9VS59 B4PK68 B3NFE2 Q2M142 B4ML70 B4LDW6 A0A0M4E8R9 Q16MV0 S4UAH5 A0A1L3N221 A0A0A1WHV1 A0A1A9WCW0 B3M5R2 A0A3B0J618 A0A1W4UNU3 A0A1A9VMU2 A0A1A9Z8W3 D3TSF1 A0A0M9A5Y4 W8BNC5 A0A034WPL1 A0A0C9QIA2 A0A0K8VQS4 A0A2A3EBB0 V9IJI1 A0A088AHT4 E5L9J4 A0A1A9Y9Y9 A0A1B0BQ76 A0A154PTI5 K7IUT5 A0A1B6L1T5 A0A1B6GEC8 A0A1B6I263 A0A0L7RIM6 A0A1B6JQA6 A0A067R299 A0A0C9QZ52 D6WLM5 A0A2P8XZD6 A0A0T6B914 A0A1B6DP15 B4J2H1 N6UJ91 A0A232FBL3 A0A1Y1JWR6 A0A1E1X1X1 A0A0L8IF12
Pubmed
19121390
23622113
22118469
26354079
26369729
24330624
+ More
24945155 25244985 20966253 12364791 14747013 17210077 26483478 19229557 25315136 26108605 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 18327897 18066067 25180232 24947515 17550304 15632085 17510324 25830018 18057021 20353571 24495485 25348373 20978910 20075255 24845553 18362917 19820115 29403074 23537049 28648823 28004739 28503490
24945155 25244985 20966253 12364791 14747013 17210077 26483478 19229557 25315136 26108605 17994087 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 12537569 18327897 18066067 25180232 24947515 17550304 15632085 17510324 25830018 18057021 20353571 24495485 25348373 20978910 20075255 24845553 18362917 19820115 29403074 23537049 28648823 28004739 28503490
EMBL
JQ289152
AEZ56541.1
BABH01030749
BABH01030750
GAIX01012007
JAA80553.1
+ More
NWSH01005788 PCG63956.1 AGBW02012108 OWR45628.1 KQ460416 KPJ14910.1 KQ459324 KPJ01635.1 GDAI01001711 JAI15892.1 GANO01000491 JAB59380.1 KU973619 DS232242 APH10051.1 EDS38270.1 CVRI01000042 CRK95633.1 GALA01000787 JAA94065.1 GALA01000794 JAA94058.1 GFDF01009761 JAV04323.1 GALA01000724 JAA94128.1 GAPW01004581 JAC09017.1 KU973610 APH10042.1 KU973605 KU973606 APH10038.1 KU973616 APH10048.1 KU973615 APH10047.1 KU973611 APH10043.1 KU973614 APH10046.1 KU973607 KU973612 KU973620 AAAB01008807 APH10040.1 EAA04195.4 APCN01003620 KU973613 APH10045.1 JXUM01027402 JXUM01027403 JXUM01027404 KQ560789 KXJ80878.1 EU637024 KU973617 ACF49499.1 APH10049.1 AXCN02000573 KU973608 APH10041.1 GGFL01004499 MBW68677.1 GGFK01007064 MBW40385.1 GGFM01003263 MBW24014.1 KA646194 AFP60823.1 GFDL01014085 JAV20960.1 JRES01000492 KNC30812.1 CH933809 EDW17758.1 AJVK01058809 CH480817 EDW50830.1 CM000363 CM002912 EDX09560.1 KMY98080.1 AE014296 ADV37495.1 ADV37496.1 ADV37497.1 AGB94206.1 AY095189 AAF50569.1 AAM12282.1 CM000159 EDW93748.1 CH954178 EDV50484.1 CH379069 EAL30734.1 CH963847 EDW73128.2 CH940647 EDW68989.1 CP012525 ALC43328.1 KU973618 CH477848 APH10050.1 EAT35663.1 JX193853 AGI44629.1 KU973621 APH10053.1 GBXI01016197 GBXI01009315 GBXI01004652 JAC98094.1 JAD04977.1 JAD09640.1 CH902618 EDV40696.1 KPU78698.1 KPU78699.1 OUUW01000002 SPP77175.1 CCAG010020666 EZ424353 ADD20629.1 KQ435727 KOX77905.1 GAMC01011684 JAB94871.1 GAKP01002705 JAC56247.1 GBYB01003254 JAG73021.1 GDHF01020519 GDHF01011374 JAI31795.1 JAI40940.1 KZ288310 PBC28582.1 JR050267 AEY61215.1 HQ383975 ADQ64468.1 JXJN01018483 KQ435119 KZC14744.1 AAZX01002635 GEBQ01022294 JAT17683.1 GECZ01011997 GECZ01008985 JAS57772.1 JAS60784.1 GECU01026693 GECU01025064 GECU01001999 GECU01000803 JAS81013.1 JAS82642.1 JAT05708.1 JAT06904.1 KQ414583 KOC70715.1 GECU01024818 GECU01006282 JAS82888.1 JAT01425.1 KK853069 KDR11818.1 GBYB01009079 JAG78846.1 KQ971343 EFA04133.1 PYGN01001127 PSN37365.1 LJIG01009065 KRT83825.1 GEDC01009888 JAS27410.1 CH916366 EDV97056.1 APGK01032729 KB740848 KB631868 ENN78702.1 ERL86828.1 NNAY01000550 OXU27727.1 GEZM01101887 GEZM01101886 JAV52230.1 GFAC01005925 JAT93263.1 KQ415861 KOG00018.1
NWSH01005788 PCG63956.1 AGBW02012108 OWR45628.1 KQ460416 KPJ14910.1 KQ459324 KPJ01635.1 GDAI01001711 JAI15892.1 GANO01000491 JAB59380.1 KU973619 DS232242 APH10051.1 EDS38270.1 CVRI01000042 CRK95633.1 GALA01000787 JAA94065.1 GALA01000794 JAA94058.1 GFDF01009761 JAV04323.1 GALA01000724 JAA94128.1 GAPW01004581 JAC09017.1 KU973610 APH10042.1 KU973605 KU973606 APH10038.1 KU973616 APH10048.1 KU973615 APH10047.1 KU973611 APH10043.1 KU973614 APH10046.1 KU973607 KU973612 KU973620 AAAB01008807 APH10040.1 EAA04195.4 APCN01003620 KU973613 APH10045.1 JXUM01027402 JXUM01027403 JXUM01027404 KQ560789 KXJ80878.1 EU637024 KU973617 ACF49499.1 APH10049.1 AXCN02000573 KU973608 APH10041.1 GGFL01004499 MBW68677.1 GGFK01007064 MBW40385.1 GGFM01003263 MBW24014.1 KA646194 AFP60823.1 GFDL01014085 JAV20960.1 JRES01000492 KNC30812.1 CH933809 EDW17758.1 AJVK01058809 CH480817 EDW50830.1 CM000363 CM002912 EDX09560.1 KMY98080.1 AE014296 ADV37495.1 ADV37496.1 ADV37497.1 AGB94206.1 AY095189 AAF50569.1 AAM12282.1 CM000159 EDW93748.1 CH954178 EDV50484.1 CH379069 EAL30734.1 CH963847 EDW73128.2 CH940647 EDW68989.1 CP012525 ALC43328.1 KU973618 CH477848 APH10050.1 EAT35663.1 JX193853 AGI44629.1 KU973621 APH10053.1 GBXI01016197 GBXI01009315 GBXI01004652 JAC98094.1 JAD04977.1 JAD09640.1 CH902618 EDV40696.1 KPU78698.1 KPU78699.1 OUUW01000002 SPP77175.1 CCAG010020666 EZ424353 ADD20629.1 KQ435727 KOX77905.1 GAMC01011684 JAB94871.1 GAKP01002705 JAC56247.1 GBYB01003254 JAG73021.1 GDHF01020519 GDHF01011374 JAI31795.1 JAI40940.1 KZ288310 PBC28582.1 JR050267 AEY61215.1 HQ383975 ADQ64468.1 JXJN01018483 KQ435119 KZC14744.1 AAZX01002635 GEBQ01022294 JAT17683.1 GECZ01011997 GECZ01008985 JAS57772.1 JAS60784.1 GECU01026693 GECU01025064 GECU01001999 GECU01000803 JAS81013.1 JAS82642.1 JAT05708.1 JAT06904.1 KQ414583 KOC70715.1 GECU01024818 GECU01006282 JAS82888.1 JAT01425.1 KK853069 KDR11818.1 GBYB01009079 JAG78846.1 KQ971343 EFA04133.1 PYGN01001127 PSN37365.1 LJIG01009065 KRT83825.1 GEDC01009888 JAS27410.1 CH916366 EDV97056.1 APGK01032729 KB740848 KB631868 ENN78702.1 ERL86828.1 NNAY01000550 OXU27727.1 GEZM01101887 GEZM01101886 JAV52230.1 GFAC01005925 JAT93263.1 KQ415861 KOG00018.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000002320
+ More
UP000183832 UP000075885 UP000075920 UP000076408 UP000075900 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000069940 UP000249989 UP000075886 UP000075884 UP000069272 UP000095301 UP000075880 UP000095300 UP000037069 UP000009192 UP000092462 UP000001292 UP000000304 UP000000803 UP000002282 UP000008711 UP000001819 UP000007798 UP000008792 UP000092553 UP000008820 UP000091820 UP000007801 UP000268350 UP000192221 UP000078200 UP000092445 UP000092444 UP000053105 UP000242457 UP000005203 UP000092443 UP000092460 UP000076502 UP000002358 UP000053825 UP000027135 UP000007266 UP000245037 UP000001070 UP000019118 UP000030742 UP000215335 UP000053454
UP000183832 UP000075885 UP000075920 UP000076408 UP000075900 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000069940 UP000249989 UP000075886 UP000075884 UP000069272 UP000095301 UP000075880 UP000095300 UP000037069 UP000009192 UP000092462 UP000001292 UP000000304 UP000000803 UP000002282 UP000008711 UP000001819 UP000007798 UP000008792 UP000092553 UP000008820 UP000091820 UP000007801 UP000268350 UP000192221 UP000078200 UP000092445 UP000092444 UP000053105 UP000242457 UP000005203 UP000092443 UP000092460 UP000076502 UP000002358 UP000053825 UP000027135 UP000007266 UP000245037 UP000001070 UP000019118 UP000030742 UP000215335 UP000053454
Interpro
IPR024132
Akirin
ProteinModelPortal
H6WS69
H9JT47
S4NXV2
A0A2A4IWW2
A0A212EVV2
A0A194RCA2
+ More
A0A194Q9G3 A0A0K8TNC4 U5EWM8 B0X0M8 A0A1J1IB17 T1DF49 T1E2E5 A0A1L8DCY5 T1E2G9 A0A182PIV6 A0A182WI31 A0A023EJ88 A0A1L3N235 A0A182Y884 A0A1L3N228 A0A182R6Q1 A0A182VLD0 A0A182L3Z9 A0A182X279 Q7QIE8 A0A182I6D9 A0A182H6N2 C3RX28 A0A182QLK0 A0A1L3N225 A0A182N7F5 A0A2M4CTL6 A0A2M4AHW6 A0A182FFJ8 A0A1I8MWN9 A0A2M3Z691 T1PBY3 A0A1Q3F078 A0A182J5N6 A0A1I8NMV6 A0A0L0CF21 B4KYP4 A0A1B0DFL2 B4HV59 B4QKD7 M9MRW4 Q9VS59 B4PK68 B3NFE2 Q2M142 B4ML70 B4LDW6 A0A0M4E8R9 Q16MV0 S4UAH5 A0A1L3N221 A0A0A1WHV1 A0A1A9WCW0 B3M5R2 A0A3B0J618 A0A1W4UNU3 A0A1A9VMU2 A0A1A9Z8W3 D3TSF1 A0A0M9A5Y4 W8BNC5 A0A034WPL1 A0A0C9QIA2 A0A0K8VQS4 A0A2A3EBB0 V9IJI1 A0A088AHT4 E5L9J4 A0A1A9Y9Y9 A0A1B0BQ76 A0A154PTI5 K7IUT5 A0A1B6L1T5 A0A1B6GEC8 A0A1B6I263 A0A0L7RIM6 A0A1B6JQA6 A0A067R299 A0A0C9QZ52 D6WLM5 A0A2P8XZD6 A0A0T6B914 A0A1B6DP15 B4J2H1 N6UJ91 A0A232FBL3 A0A1Y1JWR6 A0A1E1X1X1 A0A0L8IF12
A0A194Q9G3 A0A0K8TNC4 U5EWM8 B0X0M8 A0A1J1IB17 T1DF49 T1E2E5 A0A1L8DCY5 T1E2G9 A0A182PIV6 A0A182WI31 A0A023EJ88 A0A1L3N235 A0A182Y884 A0A1L3N228 A0A182R6Q1 A0A182VLD0 A0A182L3Z9 A0A182X279 Q7QIE8 A0A182I6D9 A0A182H6N2 C3RX28 A0A182QLK0 A0A1L3N225 A0A182N7F5 A0A2M4CTL6 A0A2M4AHW6 A0A182FFJ8 A0A1I8MWN9 A0A2M3Z691 T1PBY3 A0A1Q3F078 A0A182J5N6 A0A1I8NMV6 A0A0L0CF21 B4KYP4 A0A1B0DFL2 B4HV59 B4QKD7 M9MRW4 Q9VS59 B4PK68 B3NFE2 Q2M142 B4ML70 B4LDW6 A0A0M4E8R9 Q16MV0 S4UAH5 A0A1L3N221 A0A0A1WHV1 A0A1A9WCW0 B3M5R2 A0A3B0J618 A0A1W4UNU3 A0A1A9VMU2 A0A1A9Z8W3 D3TSF1 A0A0M9A5Y4 W8BNC5 A0A034WPL1 A0A0C9QIA2 A0A0K8VQS4 A0A2A3EBB0 V9IJI1 A0A088AHT4 E5L9J4 A0A1A9Y9Y9 A0A1B0BQ76 A0A154PTI5 K7IUT5 A0A1B6L1T5 A0A1B6GEC8 A0A1B6I263 A0A0L7RIM6 A0A1B6JQA6 A0A067R299 A0A0C9QZ52 D6WLM5 A0A2P8XZD6 A0A0T6B914 A0A1B6DP15 B4J2H1 N6UJ91 A0A232FBL3 A0A1Y1JWR6 A0A1E1X1X1 A0A0L8IF12
Ontologies
GO
PANTHER
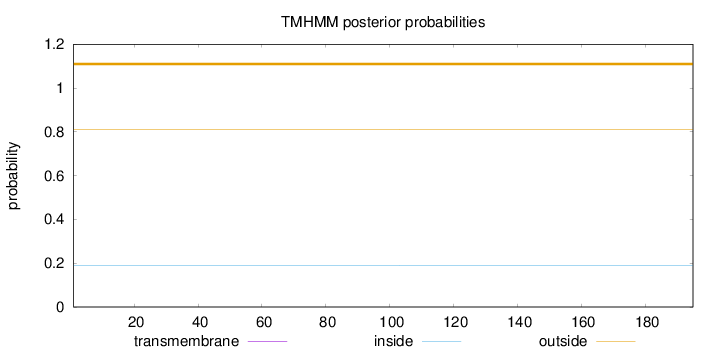
Topology
Subcellular location
Nucleus
Length:
195
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.18840
outside
1 - 195
Population Genetic Test Statistics
Pi
157.466322
Theta
169.024178
Tajima's D
-0.459202
CLR
0.421345
CSRT
0.250337483125844
Interpretation
Uncertain
