Gene
KWMTBOMO12852
Pre Gene Modal
BGIBMGA010729
Annotation
retroelement_polyprotein_[Glyptapanteles_flavicoxis]
Location in the cell
Nuclear Reliability : 2.524
Sequence
CDS
ATGGGTGACTGTGACACTGAATCTCGCTATTGTACTACTTTGACAGTTGAGTTTCCCGAAATTACGCTAGAAATAGACTTTCAAATTGTTAGTGACAAGTTCATGAATACCCCAGTCATAATCGGTACTGACGTTTTGAATCGGGAAGGTGTCACGTATATTAGGAAAAAGGGCATACAGCGCTTAACTCGGGTAGAAAAGGTTTTTACTGCGGAGACAACTGAACTTAATTCAATCCGTACCGTGAATACTTCATTAACAGGTGAAGAAAAAAATAAATTATTAGCTTTGGTACGAAAATTTTCAAAATACCTTATAACTGGTACCGCGGCCAGTACTGTTACAACTGGATCCATGCAAATTCGACTCAGCAATGAAACTCCTATTGTTTACAGACCTTACAAATTATCATTTAGCGAAAAACTTCGCGTTAGAGAAATTATTAAGGATCTTCTTGATAAAGGTGTGATTAGGGAGTCTGAATCGGAGTTCTCGAGTCCGATATTGCTAGTTAAGAAAAAGGACGGCAGTGACAGGATGTGCGTGGACTTTCGGGCTCTTAATGCGAATACTGTGAAGGATCGCTATCCATTGCCGTTAATAGACGATCACATTGACCGGTTAGGTAGCGCAAAATATTTTACAAGCCTCGATATGGCCACTGGTTTCCACCAGATTCCTTTAGCAAACGACTCCATTCACAAAACCGGCTTTGTGACACCCGAAGGTCATTTTGAATACCTTAAAATGCCCTACGGGTTAGCTAATGCCCCGGTTGTTTATCAACGTATTATAACTAATACTCTGAAAGCCTTTAGTGAGACAGGGAGGGCTTTAATCTATATCGATGATGTTTTAATACCTAGTCAGACCATTGATGAAGGCCTAAATACTCTGCGTGAAATCTTTGAAACACTAACTAAGGCGGGCTTCTCAATTAATTTAAAAAAATGCAAGTTTCTTGCTAACGAAATTGAATATTTGGGCAGAACTATCAGCAATGGTCAGGTTAGGCCGAGTACGTATAAAGTAGAAGCATTGACAAAAGCCCCGAAGCCTGGTAATGTTAAACAAGCTAGACAATTTTTAGGTCTAGCGGGTTATTTTCGGCGCTATATTCCTGCATATTCGTTGCAGACAGCTTGTATTTCTAATCTGTTTCGTAAAGGTGTTGAGTTTAAATGGGGTGATGAACAGGAACAAGTACGCCAGTATATAATTACGCGTTTAACCAGTGAACCTGTCCTCAATATATTTAATCCATCATTATTAACAGAAGTACACACTGATGCTTCTTCTCGAGGGTATGGAGCTGTTTTACTTCAGACTCATGGGAATAATAATAAACGCGTGGTGGGATATTTTAGTAAAGTTACCCAAGGCGCCGAACCACGCTATCACTCCTATGAGCTGGAGACACTCGCTGTGGTCAAGGCTCTGCAGCATTTCAGGCACTACCTCATAGGTATACATTTCAAGGTAGTTACAGACTGCAACGCTTTGAAGCTTACCCAACGTAAAAAAGATTTACTACCGAGAGTCGCTCGGTGGTGGATCTACTTACAAGATTATGATTTTTCGTTAGAGTATCGTAAGGGCGCTGTGCTATCTCATGCCGACTATCTAAGTCGTAATCCTATAAATGTTTGTGAAATTGGTAGACCGCATAACTGGGCACAGATAGCACAGAGTGCTGATGAAGAAACTCGGAATTTAATACAAAAGTTACAAAACGGTGAATTAGATGAGACACGCTATGTAACTAAAAATGATGTTCTTTATTATAAATTTGATTCTGTTGGCCAACCTAGTAAATTATTATGTTATATTCCAAGAGGACATCGCCTCAGTTTGTTACGAATCTTTCATGATGAGCATCATCATATTGGAGTAGATAAGACGCTTGATCTAATATTAAACCATTTTTGGTTTCCTAGTCTAAGACAATTTGTTCGTAAATATGTACGCCATTGTATAGTATGTCTTTCACACAAAAAAATACCACGTCAACCCTTGCAGCCGATCGAGTCGTGGAAGAAACCGGACGTACCCTTTGACACGGTACATAGTGATGTCTTGGGTCCTCTTCCCGAATCGAACGGTTACAAATTTGTTGTCTTGATAATTGACGCTTATAGTAAGTTCTGTCTACTAAATCCTATGCGTAAGCAGGATGCAACAGAGTTAAAATTAGTATTTGAAAACGCTGTATCTTTATTTGGTGCACCTAGGCAGATTGTAACCGATAGGGGTAGAATGTACGAAAATAGATCATTTAGACAATGGGTATCAGACGTAGGTAGTAATTTATTCTTTATAACGCCAGAGATGCACCAGAGTAACGGTCAGGCTGAGAGATATTGCAGAACAGTTCTTAATATGATTCGTGTGGAGGTAAATTTCAGAAACGAAAATTGGTCAGAAGTTTTGTGGAAACTGCAGCTCACCCTGAACATTACGAAACAAAAGTCCACTCAATATTCACCTTTATATTTGTTAATTGGTAGCGATGCTGTGACGCCCGTCATTCGTTCCGTTGTACGCGATGTGGCTCTAGAAGGAACAAGTGCCAACAGAGAAACCTTACGCGAGCTTGCCCGACAAAGAGCTTCTCAACTGCTGGACAAAAACCGAAAACAGCAGGACACTCGAGTAAATGAACGGCGCAAACCCCCGCGTACTTTCCATGAAAACGACGTGGTCTTCGTGCATAAGAATAGCCAATCTGTTGGAAAGCTGGATTCTGGTATGCGTGGCCCGTACAAAATAGTGCGTGCGCTGCCACATGGACGTTATGAACTGAAGCTTCTTTCTGGCTCACTTGGGAAGACCACGAAAGCTGCTGCCGAATTTATAAGTGATGAGAATAGCTCTGAGGAAGGTATTGTTGCCGGTCCATCACACCACCATGACGACATAGTACCTCTAGACGGACCTATTAATGCGGGCGAGGACGCTCCGCCGTCAGGAGAGGCCGTGTTAGAAGATGATGATGACATTGGAGGCGTGGCCTAA
Protein
MGDCDTESRYCTTLTVEFPEITLEIDFQIVSDKFMNTPVIIGTDVLNREGVTYIRKKGIQRLTRVEKVFTAETTELNSIRTVNTSLTGEEKNKLLALVRKFSKYLITGTAASTVTTGSMQIRLSNETPIVYRPYKLSFSEKLRVREIIKDLLDKGVIRESESEFSSPILLVKKKDGSDRMCVDFRALNANTVKDRYPLPLIDDHIDRLGSAKYFTSLDMATGFHQIPLANDSIHKTGFVTPEGHFEYLKMPYGLANAPVVYQRIITNTLKAFSETGRALIYIDDVLIPSQTIDEGLNTLREIFETLTKAGFSINLKKCKFLANEIEYLGRTISNGQVRPSTYKVEALTKAPKPGNVKQARQFLGLAGYFRRYIPAYSLQTACISNLFRKGVEFKWGDEQEQVRQYIITRLTSEPVLNIFNPSLLTEVHTDASSRGYGAVLLQTHGNNNKRVVGYFSKVTQGAEPRYHSYELETLAVVKALQHFRHYLIGIHFKVVTDCNALKLTQRKKDLLPRVARWWIYLQDYDFSLEYRKGAVLSHADYLSRNPINVCEIGRPHNWAQIAQSADEETRNLIQKLQNGELDETRYVTKNDVLYYKFDSVGQPSKLLCYIPRGHRLSLLRIFHDEHHHIGVDKTLDLILNHFWFPSLRQFVRKYVRHCIVCLSHKKIPRQPLQPIESWKKPDVPFDTVHSDVLGPLPESNGYKFVVLIIDAYSKFCLLNPMRKQDATELKLVFENAVSLFGAPRQIVTDRGRMYENRSFRQWVSDVGSNLFFITPEMHQSNGQAERYCRTVLNMIRVEVNFRNENWSEVLWKLQLTLNITKQKSTQYSPLYLLIGSDAVTPVIRSVVRDVALEGTSANRETLRELARQRASQLLDKNRKQQDTRVNERRKPPRTFHENDVVFVHKNSQSVGKLDSGMRGPYKIVRALPHGRYELKLLSGSLGKTTKAAAEFISDENSSEEGIVAGPSHHHDDIVPLDGPINAGEDAPPSGEAVLEDDDDIGGVA
Summary
Uniprot
A0A437AUZ6
A0A0A9X1Z8
A0A146M2J2
B7S8P8
A0A0J7KEC9
A0A0C9RXL6
+ More
A0A0A9WJJ8 A0A0A9Z954 A0A139WJA1 A0A139WIQ0 A0A3S2L135 D2A5M1 A0A194R1L3 A0A1Y1NG42 A0A1Y1KFN9 A0A1Y1KFM9 A0A1Y1NAV9 J9JJ45 A0A147BLC5 A0A0J7NF11 A0A2S2P032 A0A0J7KLK4 A0A1Y1MFA9 A0A139WIB9 A0A0J7K9X8 A0A2S2PGQ9 X1X5H2 X1WPP0 A0A2S2NFQ4 A0A2H8TRV6 J9LBS9 X1XM84 A0A1W7R6G2 A0A0J7K890 A0A1Y1MFE0 W8C8Q1 A0A2S2NKJ2 X1XD76 X1XU14 Q24310 W8B0V9 A0A2L2Y402 A0A2L2Y1I8 A0A2L2Y1Z9 A0A034WRX5 A0A1Y1NEJ5 J9JQ65 A0A0A1XB16 A0A0A1XFE9 A0A023EZG2 A0A034VLR3 A0A023EYH9 J9JZ77 A0A2S2PFS2 A0A2S2QVZ0 A0A023EY26 A0A1W7R6L8 A0A034VPR8 A0A034VKV9 W8APB2 A0A224XHR1 A0A1W7R6R7 A0A2S2QIS3 A0A034W795 X1XU22 A0A1Y1LN89 A0A0A1WKC8 A0A1W7R6G1 A0A2S2R3T0 A0A139WCG2 A0A139WCC5 A0A2A4J7U9 T1PMR9 X1X261 A0A2S2PMX3 A0A1W7R6H1 Q94885 A0A1Z5L674 A0A147BTY1 L7LV55 A0A131XQ77 A0A034VAM6 A0A2M4AI73 A0A023F013 A0A023EYF0 A0A0A9YEP8 A0A2M4AEU2 L7MCS8 L7LY95 J9K544
A0A0A9WJJ8 A0A0A9Z954 A0A139WJA1 A0A139WIQ0 A0A3S2L135 D2A5M1 A0A194R1L3 A0A1Y1NG42 A0A1Y1KFN9 A0A1Y1KFM9 A0A1Y1NAV9 J9JJ45 A0A147BLC5 A0A0J7NF11 A0A2S2P032 A0A0J7KLK4 A0A1Y1MFA9 A0A139WIB9 A0A0J7K9X8 A0A2S2PGQ9 X1X5H2 X1WPP0 A0A2S2NFQ4 A0A2H8TRV6 J9LBS9 X1XM84 A0A1W7R6G2 A0A0J7K890 A0A1Y1MFE0 W8C8Q1 A0A2S2NKJ2 X1XD76 X1XU14 Q24310 W8B0V9 A0A2L2Y402 A0A2L2Y1I8 A0A2L2Y1Z9 A0A034WRX5 A0A1Y1NEJ5 J9JQ65 A0A0A1XB16 A0A0A1XFE9 A0A023EZG2 A0A034VLR3 A0A023EYH9 J9JZ77 A0A2S2PFS2 A0A2S2QVZ0 A0A023EY26 A0A1W7R6L8 A0A034VPR8 A0A034VKV9 W8APB2 A0A224XHR1 A0A1W7R6R7 A0A2S2QIS3 A0A034W795 X1XU22 A0A1Y1LN89 A0A0A1WKC8 A0A1W7R6G1 A0A2S2R3T0 A0A139WCG2 A0A139WCC5 A0A2A4J7U9 T1PMR9 X1X261 A0A2S2PMX3 A0A1W7R6H1 Q94885 A0A1Z5L674 A0A147BTY1 L7LV55 A0A131XQ77 A0A034VAM6 A0A2M4AI73 A0A023F013 A0A023EYF0 A0A0A9YEP8 A0A2M4AEU2 L7MCS8 L7LY95 J9K544
Pubmed
EMBL
RSAL01000399
RVE41947.1
GBHO01030786
JAG12818.1
GDHC01004980
JAQ13649.1
+ More
EF710649 ACE75273.1 LBMM01008871 KMQ88569.1 GBYB01013920 JAG83687.1 GBHO01036023 GBHO01032007 JAG07581.1 JAG11597.1 GBHO01040010 GBHO01001812 JAG03594.1 JAG41792.1 KQ971338 KYB27887.1 KYB27888.1 RSAL01000274 RVE43148.1 KQ971345 EFA05054.2 KQ460883 KPJ11409.1 GEZM01007614 GEZM01007613 GEZM01007612 JAV95056.1 GEZM01089268 GEZM01089261 JAV57577.1 GEZM01089269 GEZM01089265 JAV57567.1 GEZM01007616 GEZM01007615 GEZM01007611 JAV95051.1 ABLF02008401 ABLF02008404 GEGO01004299 JAR91105.1 LBMM01005919 KMQ91095.1 GGMR01009949 MBY22568.1 KMQ91096.1 GEZM01033083 JAV84424.1 KQ971339 KYB27733.1 LBMM01010976 KMQ87102.1 GGMR01016004 MBY28623.1 ABLF02004401 ABLF02013544 ABLF02013550 ABLF02033943 ABLF02042737 GGMR01003388 MBY16007.1 GFXV01005120 MBW16925.1 ABLF02041876 ABLF02004526 ABLF02054969 GEHC01000888 JAV46757.1 LBMM01011782 KMQ86633.1 GEZM01034557 JAV83728.1 GAMC01007326 JAB99229.1 GGMR01005091 MBY17710.1 ABLF02022032 ABLF02003120 ABLF02004015 ABLF02057179 ABLF02066208 X14037 CAA32198.1 GAMC01015907 JAB90648.1 IAAA01004057 LAA02040.1 IAAA01004058 LAA02042.1 IAAA01004056 LAA02037.1 GAKP01001563 JAC57389.1 GEZM01005427 JAV96179.1 ABLF02005846 GBXI01005773 JAD08519.1 GBXI01004632 JAD09660.1 GBBI01004551 JAC14161.1 GAKP01016237 JAC42715.1 GBBI01004550 JAC14162.1 ABLF02014660 ABLF02042774 GGMR01015682 MBY28301.1 GGMS01012726 MBY81929.1 GBBI01004477 JAC14235.1 GEHC01000930 JAV46715.1 GAKP01014483 JAC44469.1 GAKP01016235 JAC42717.1 GAMC01020207 JAB86348.1 GFTR01008753 JAW07673.1 GEHC01000880 JAV46765.1 GGMS01008442 MBY77645.1 GAKP01008942 JAC50010.1 ABLF02017401 GEZM01051217 GEZM01051215 JAV75083.1 GBXI01015151 JAC99140.1 GEHC01000891 JAV46754.1 GGMS01015422 MBY84625.1 KQ971370 KYB25610.1 KYB25609.1 NWSH01002797 PCG67520.1 KA649989 AFP64618.1 ABLF02005127 ABLF02005129 ABLF02007401 GGMR01018192 MBY30811.1 GEHC01000876 JAV46769.1 X95908 CAA65152.1 GFJQ02004121 JAW02849.1 GEGO01001472 JAR93932.1 GACK01009547 JAA55487.1 GEFM01006999 JAP68797.1 GAKP01019383 JAC39569.1 GGFK01007168 MBW40489.1 GBBI01004613 JAC14099.1 GBBI01004549 JAC14163.1 GBHO01015604 JAG28000.1 GGFK01005990 MBW39311.1 GACK01004001 JAA61033.1 GACK01009195 JAA55839.1 ABLF02023666
EF710649 ACE75273.1 LBMM01008871 KMQ88569.1 GBYB01013920 JAG83687.1 GBHO01036023 GBHO01032007 JAG07581.1 JAG11597.1 GBHO01040010 GBHO01001812 JAG03594.1 JAG41792.1 KQ971338 KYB27887.1 KYB27888.1 RSAL01000274 RVE43148.1 KQ971345 EFA05054.2 KQ460883 KPJ11409.1 GEZM01007614 GEZM01007613 GEZM01007612 JAV95056.1 GEZM01089268 GEZM01089261 JAV57577.1 GEZM01089269 GEZM01089265 JAV57567.1 GEZM01007616 GEZM01007615 GEZM01007611 JAV95051.1 ABLF02008401 ABLF02008404 GEGO01004299 JAR91105.1 LBMM01005919 KMQ91095.1 GGMR01009949 MBY22568.1 KMQ91096.1 GEZM01033083 JAV84424.1 KQ971339 KYB27733.1 LBMM01010976 KMQ87102.1 GGMR01016004 MBY28623.1 ABLF02004401 ABLF02013544 ABLF02013550 ABLF02033943 ABLF02042737 GGMR01003388 MBY16007.1 GFXV01005120 MBW16925.1 ABLF02041876 ABLF02004526 ABLF02054969 GEHC01000888 JAV46757.1 LBMM01011782 KMQ86633.1 GEZM01034557 JAV83728.1 GAMC01007326 JAB99229.1 GGMR01005091 MBY17710.1 ABLF02022032 ABLF02003120 ABLF02004015 ABLF02057179 ABLF02066208 X14037 CAA32198.1 GAMC01015907 JAB90648.1 IAAA01004057 LAA02040.1 IAAA01004058 LAA02042.1 IAAA01004056 LAA02037.1 GAKP01001563 JAC57389.1 GEZM01005427 JAV96179.1 ABLF02005846 GBXI01005773 JAD08519.1 GBXI01004632 JAD09660.1 GBBI01004551 JAC14161.1 GAKP01016237 JAC42715.1 GBBI01004550 JAC14162.1 ABLF02014660 ABLF02042774 GGMR01015682 MBY28301.1 GGMS01012726 MBY81929.1 GBBI01004477 JAC14235.1 GEHC01000930 JAV46715.1 GAKP01014483 JAC44469.1 GAKP01016235 JAC42717.1 GAMC01020207 JAB86348.1 GFTR01008753 JAW07673.1 GEHC01000880 JAV46765.1 GGMS01008442 MBY77645.1 GAKP01008942 JAC50010.1 ABLF02017401 GEZM01051217 GEZM01051215 JAV75083.1 GBXI01015151 JAC99140.1 GEHC01000891 JAV46754.1 GGMS01015422 MBY84625.1 KQ971370 KYB25610.1 KYB25609.1 NWSH01002797 PCG67520.1 KA649989 AFP64618.1 ABLF02005127 ABLF02005129 ABLF02007401 GGMR01018192 MBY30811.1 GEHC01000876 JAV46769.1 X95908 CAA65152.1 GFJQ02004121 JAW02849.1 GEGO01001472 JAR93932.1 GACK01009547 JAA55487.1 GEFM01006999 JAP68797.1 GAKP01019383 JAC39569.1 GGFK01007168 MBW40489.1 GBBI01004613 JAC14099.1 GBBI01004549 JAC14163.1 GBHO01015604 JAG28000.1 GGFK01005990 MBW39311.1 GACK01004001 JAA61033.1 GACK01009195 JAA55839.1 ABLF02023666
Proteomes
Pfam
Interpro
IPR000477
RT_dom
+ More
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR001878 Znf_CCHC
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR018061 Retropepsins
IPR021109 Peptidase_aspartic_dom_sf
IPR036875 Znf_CCHC_sf
IPR041373 RT_RNaseH
IPR034122 Retropepsin-like_bacterial
IPR025836 Zn_knuckle_CX2CX4HX4C
IPR028002 Myb_DNA-bind_5
IPR036361 SAP_dom_sf
IPR003034 SAP_dom
IPR005162 Retrotrans_gag_dom
IPR032071 DUF4806
IPR024650 Peptidase_A2B
IPR019103 Peptidase_aspartic_DDI1-type
IPR004955 Baculovirus_Gp64
IPR006579 Pre_C2HC_dom
IPR011764 Biotin_carboxylation_dom
IPR041588 Integrase_H2C2
IPR036397 RNaseH_sf
IPR012337 RNaseH-like_sf
IPR001584 Integrase_cat-core
IPR041577 RT_RNaseH_2
IPR001878 Znf_CCHC
IPR001969 Aspartic_peptidase_AS
IPR001995 Peptidase_A2_cat
IPR018061 Retropepsins
IPR021109 Peptidase_aspartic_dom_sf
IPR036875 Znf_CCHC_sf
IPR041373 RT_RNaseH
IPR034122 Retropepsin-like_bacterial
IPR025836 Zn_knuckle_CX2CX4HX4C
IPR028002 Myb_DNA-bind_5
IPR036361 SAP_dom_sf
IPR003034 SAP_dom
IPR005162 Retrotrans_gag_dom
IPR032071 DUF4806
IPR024650 Peptidase_A2B
IPR019103 Peptidase_aspartic_DDI1-type
IPR004955 Baculovirus_Gp64
IPR006579 Pre_C2HC_dom
IPR011764 Biotin_carboxylation_dom
Gene 3D
ProteinModelPortal
A0A437AUZ6
A0A0A9X1Z8
A0A146M2J2
B7S8P8
A0A0J7KEC9
A0A0C9RXL6
+ More
A0A0A9WJJ8 A0A0A9Z954 A0A139WJA1 A0A139WIQ0 A0A3S2L135 D2A5M1 A0A194R1L3 A0A1Y1NG42 A0A1Y1KFN9 A0A1Y1KFM9 A0A1Y1NAV9 J9JJ45 A0A147BLC5 A0A0J7NF11 A0A2S2P032 A0A0J7KLK4 A0A1Y1MFA9 A0A139WIB9 A0A0J7K9X8 A0A2S2PGQ9 X1X5H2 X1WPP0 A0A2S2NFQ4 A0A2H8TRV6 J9LBS9 X1XM84 A0A1W7R6G2 A0A0J7K890 A0A1Y1MFE0 W8C8Q1 A0A2S2NKJ2 X1XD76 X1XU14 Q24310 W8B0V9 A0A2L2Y402 A0A2L2Y1I8 A0A2L2Y1Z9 A0A034WRX5 A0A1Y1NEJ5 J9JQ65 A0A0A1XB16 A0A0A1XFE9 A0A023EZG2 A0A034VLR3 A0A023EYH9 J9JZ77 A0A2S2PFS2 A0A2S2QVZ0 A0A023EY26 A0A1W7R6L8 A0A034VPR8 A0A034VKV9 W8APB2 A0A224XHR1 A0A1W7R6R7 A0A2S2QIS3 A0A034W795 X1XU22 A0A1Y1LN89 A0A0A1WKC8 A0A1W7R6G1 A0A2S2R3T0 A0A139WCG2 A0A139WCC5 A0A2A4J7U9 T1PMR9 X1X261 A0A2S2PMX3 A0A1W7R6H1 Q94885 A0A1Z5L674 A0A147BTY1 L7LV55 A0A131XQ77 A0A034VAM6 A0A2M4AI73 A0A023F013 A0A023EYF0 A0A0A9YEP8 A0A2M4AEU2 L7MCS8 L7LY95 J9K544
A0A0A9WJJ8 A0A0A9Z954 A0A139WJA1 A0A139WIQ0 A0A3S2L135 D2A5M1 A0A194R1L3 A0A1Y1NG42 A0A1Y1KFN9 A0A1Y1KFM9 A0A1Y1NAV9 J9JJ45 A0A147BLC5 A0A0J7NF11 A0A2S2P032 A0A0J7KLK4 A0A1Y1MFA9 A0A139WIB9 A0A0J7K9X8 A0A2S2PGQ9 X1X5H2 X1WPP0 A0A2S2NFQ4 A0A2H8TRV6 J9LBS9 X1XM84 A0A1W7R6G2 A0A0J7K890 A0A1Y1MFE0 W8C8Q1 A0A2S2NKJ2 X1XD76 X1XU14 Q24310 W8B0V9 A0A2L2Y402 A0A2L2Y1I8 A0A2L2Y1Z9 A0A034WRX5 A0A1Y1NEJ5 J9JQ65 A0A0A1XB16 A0A0A1XFE9 A0A023EZG2 A0A034VLR3 A0A023EYH9 J9JZ77 A0A2S2PFS2 A0A2S2QVZ0 A0A023EY26 A0A1W7R6L8 A0A034VPR8 A0A034VKV9 W8APB2 A0A224XHR1 A0A1W7R6R7 A0A2S2QIS3 A0A034W795 X1XU22 A0A1Y1LN89 A0A0A1WKC8 A0A1W7R6G1 A0A2S2R3T0 A0A139WCG2 A0A139WCC5 A0A2A4J7U9 T1PMR9 X1X261 A0A2S2PMX3 A0A1W7R6H1 Q94885 A0A1Z5L674 A0A147BTY1 L7LV55 A0A131XQ77 A0A034VAM6 A0A2M4AI73 A0A023F013 A0A023EYF0 A0A0A9YEP8 A0A2M4AEU2 L7MCS8 L7LY95 J9K544
PDB
4OL8
E-value=2.37135e-77,
Score=739
Ontologies
GO
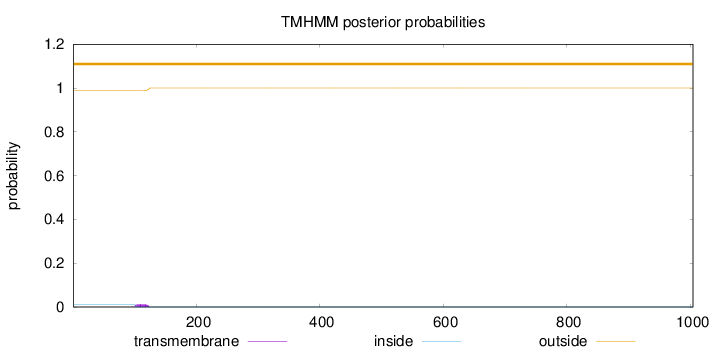
Topology
Length:
1004
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.253340000000001
Exp number, first 60 AAs:
0.00117
Total prob of N-in:
0.01195
outside
1 - 1004
Population Genetic Test Statistics
Pi
2.530243
Theta
157.463951
Tajima's D
0
CLR
0
CSRT
0.371181440927954
Interpretation
Uncertain
