Gene
KWMTBOMO12849 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012738
Annotation
PREDICTED:_peroxidase-like_isoform_X3_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.603 Mitochondrial Reliability : 1.306
Sequence
CDS
ATGGCTTATCAAAAATCATCCCTAAAGCAACAACTCTCCAAAACTTCAACTACATCAAGAGATGGTGTATTATCGTCTTCGTTTGGATACAGAGATGCGTACAATCCTGAAATACTACCACAAGTTTCGCTCGAGTACCCGTTCGTTTTGCGATGGCTCCACAGCGTCCAGGAAGGCACCCTTAAAATGTACGACACAAAGGGATATTATCTCAAGCAGTATCCGATAGTGAACCTCACTTTGAGAACAGGCTTCTTCGATGTGGACAATAACATAGATTATATAACACAAGGAGCTTTCAGACAAGGCAGTGCTAATATTGACCATACAGCTGATCCTGATATCGTTGATATCGGCTTGGGTCCTCACCAAAGAGCATCGGATCTGATGACAAACGACATCTCAAAGAACAGATATTTTGGATTCCAGCCTTACGTTAAATACCACGAATTTTTTTTCGGTAAAACGATCAAAAAATGGGGAGACCTGGAAGGAGTCATTGACCCCGAGAGGATCGAAATACTCAGAGATTTGTACGAAGATGTTGAAGACATGGACCTCTTGGCCGGTATTTGGGTTGAGAGGCGCATTAAAGGTGGTTTCGTGCGCCAACCTTCTACTGCTTGGTAG
Protein
MAYQKSSLKQQLSKTSTTSRDGVLSSSFGYRDAYNPEILPQVSLEYPFVLRWLHSVQEGTLKMYDTKGYYLKQYPIVNLTLRTGFFDVDNNIDYITQGAFRQGSANIDHTADPDIVDIGLGPHQRASDLMTNDISKNRYFGFQPYVKYHEFFFGKTIKKWGDLEGVIDPERIEILRDLYEDVEDMDLLAGIWVERRIKGGFVRQPSTAW
Summary
Uniprot
A0A2H1WGR9
A0A2A4IUC0
A0A2W1BYE5
A0A194Q9H2
A0A194Q7Y0
I4DS54
+ More
A0A194RBC8 A0A194QDV6 I4DJ10 A0A3S2NLC3 A0A194RAP6 I4DN32 A0A212ETD1 A0A2A4JI21 A0A2H1W8U7 A0A088MGW5 A0A2W1BT77 A0A2H1W4U4 A0A2H1W9U4 A0A2W1BT67 A0A2A4J7D3 H9JT77 A0A2H1WBG2 H9ISL2 H9JT78 A0A2H1VUX0 A0A194Q3N0 H9ISL1 A0A0N0PED8 H9JQT0 A0A437AS20 A0A437ASV3 A0A3S2L850 A0A3S2NMV9 A0A2H1WAX3 A0A437AYB0 A0A0L7LKV1 A0A2W1BQ93 A0A2A4IUP3 H9IT78 A0A3S2M0V1 A0A2W1BXH1 A0A2W1BS42 A0A2H1UZX5 A0A2A4IUD5 A0A2A4JM77 A0A2A4JMB0 B4JHE0 A0A2L2Y2A3 A0A0A7LYX9 A0A194Q1N0 A0A310SND6 A0A1B6MDD3 J9KYF6 K7IYZ3 A0A232F2X4 B4KC82 A0A2M4CUH8 A0A2M4A9K3 A0A2M4DKI6 A0A2P6K367 A0A0L0CRA6 D2A572 A0A0M4F792 A0A2M4AT71 G3MHL8 B4NJ86 A0A0Q9WVU0 A0A1W4VTT3 B4IBN7 W5JPI5 A0A182G6R1 B4QWD4 B4M5J8 A0A1Y9G9T7 A0A0P4VM15 A0A336MLW0 A0A2S2QB91 B4G4K4 B5DWL8 T1I7W3 A0A1D2MQ77
A0A194RBC8 A0A194QDV6 I4DJ10 A0A3S2NLC3 A0A194RAP6 I4DN32 A0A212ETD1 A0A2A4JI21 A0A2H1W8U7 A0A088MGW5 A0A2W1BT77 A0A2H1W4U4 A0A2H1W9U4 A0A2W1BT67 A0A2A4J7D3 H9JT77 A0A2H1WBG2 H9ISL2 H9JT78 A0A2H1VUX0 A0A194Q3N0 H9ISL1 A0A0N0PED8 H9JQT0 A0A437AS20 A0A437ASV3 A0A3S2L850 A0A3S2NMV9 A0A2H1WAX3 A0A437AYB0 A0A0L7LKV1 A0A2W1BQ93 A0A2A4IUP3 H9IT78 A0A3S2M0V1 A0A2W1BXH1 A0A2W1BS42 A0A2H1UZX5 A0A2A4IUD5 A0A2A4JM77 A0A2A4JMB0 B4JHE0 A0A2L2Y2A3 A0A0A7LYX9 A0A194Q1N0 A0A310SND6 A0A1B6MDD3 J9KYF6 K7IYZ3 A0A232F2X4 B4KC82 A0A2M4CUH8 A0A2M4A9K3 A0A2M4DKI6 A0A2P6K367 A0A0L0CRA6 D2A572 A0A0M4F792 A0A2M4AT71 G3MHL8 B4NJ86 A0A0Q9WVU0 A0A1W4VTT3 B4IBN7 W5JPI5 A0A182G6R1 B4QWD4 B4M5J8 A0A1Y9G9T7 A0A0P4VM15 A0A336MLW0 A0A2S2QB91 B4G4K4 B5DWL8 T1I7W3 A0A1D2MQ77
Pubmed
EMBL
ODYU01008556
SOQ52260.1
NWSH01006580
PCG63355.1
KZ149907
PZC78257.1
+ More
KQ459324 KPJ01645.1 KPJ01647.1 AK405426 BAM20744.1 KQ460416 KPJ14922.1 KPJ01646.1 AK401278 BAM17900.1 RSAL01000046 RVE50539.1 KPJ14923.1 AK402700 BAM19322.1 AGBW02012596 OWR44740.1 NWSH01001320 PCG71707.1 ODYU01007063 SOQ49501.1 KJ995811 AIN39494.1 PZC78259.1 ODYU01006337 SOQ48101.1 ODYU01007226 SOQ49823.1 PZC78258.1 NWSH01002854 PCG67434.1 BABH01034743 BABH01034744 ODYU01007479 SOQ50286.1 BABH01013447 BABH01013448 BABH01034746 BABH01034747 ODYU01004596 SOQ44641.1 KQ459564 KPI99614.1 BABH01013455 KQ459837 KPJ19761.1 BABH01028746 BABH01028747 BABH01028748 RSAL01001640 RVE40744.1 RSAL01000844 RVE41114.1 RVE41111.1 RSAL01000231 RVE43837.1 ODYU01007325 SOQ50002.1 RSAL01000273 RVE43156.1 JTDY01000732 KOB76072.1 KZ149956 PZC76461.1 NWSH01006414 PCG63451.1 BABH01013458 BABH01013459 BABH01013460 RSAL01000083 RVE48485.1 PZC76463.1 PZC76464.1 ODYU01000046 SOQ34155.1 NWSH01006200 PCG63597.1 NWSH01001116 PCG72533.1 PCG72532.1 CH916369 EDV93847.1 IAAA01005455 IAAA01005456 LAA02302.1 KM667939 AIZ68326.1 KQ459579 KPI99456.1 KQ760645 OAD59676.1 GEBQ01006041 JAT33936.1 ABLF02014398 NNAY01001189 OXU24803.1 CH933806 EDW13691.2 GGFL01004824 MBW69002.1 GGFK01004124 MBW37445.1 GGFL01013885 MBW78063.1 MWRG01033899 PRD20757.1 JRES01000032 KNC34772.1 KQ971361 EFA05698.1 CP012526 ALC47695.1 GGFK01010643 MBW43964.1 JO841369 AEO32986.1 CH964272 EDW83879.1 KRF99908.1 CH480827 EDW44795.1 ADMH02000540 ETN66036.1 JXUM01148651 KQ570609 KXJ68331.1 CM000364 EDX12625.1 CH940652 EDW58924.2 GDKW01003332 JAI53263.1 UFQT01001180 SSX29357.1 GGMS01005259 MBY74462.1 CH479179 EDW25175.1 CM000070 EDY68158.2 ACPB03011632 LJIJ01000734 ODM94915.1
KQ459324 KPJ01645.1 KPJ01647.1 AK405426 BAM20744.1 KQ460416 KPJ14922.1 KPJ01646.1 AK401278 BAM17900.1 RSAL01000046 RVE50539.1 KPJ14923.1 AK402700 BAM19322.1 AGBW02012596 OWR44740.1 NWSH01001320 PCG71707.1 ODYU01007063 SOQ49501.1 KJ995811 AIN39494.1 PZC78259.1 ODYU01006337 SOQ48101.1 ODYU01007226 SOQ49823.1 PZC78258.1 NWSH01002854 PCG67434.1 BABH01034743 BABH01034744 ODYU01007479 SOQ50286.1 BABH01013447 BABH01013448 BABH01034746 BABH01034747 ODYU01004596 SOQ44641.1 KQ459564 KPI99614.1 BABH01013455 KQ459837 KPJ19761.1 BABH01028746 BABH01028747 BABH01028748 RSAL01001640 RVE40744.1 RSAL01000844 RVE41114.1 RVE41111.1 RSAL01000231 RVE43837.1 ODYU01007325 SOQ50002.1 RSAL01000273 RVE43156.1 JTDY01000732 KOB76072.1 KZ149956 PZC76461.1 NWSH01006414 PCG63451.1 BABH01013458 BABH01013459 BABH01013460 RSAL01000083 RVE48485.1 PZC76463.1 PZC76464.1 ODYU01000046 SOQ34155.1 NWSH01006200 PCG63597.1 NWSH01001116 PCG72533.1 PCG72532.1 CH916369 EDV93847.1 IAAA01005455 IAAA01005456 LAA02302.1 KM667939 AIZ68326.1 KQ459579 KPI99456.1 KQ760645 OAD59676.1 GEBQ01006041 JAT33936.1 ABLF02014398 NNAY01001189 OXU24803.1 CH933806 EDW13691.2 GGFL01004824 MBW69002.1 GGFK01004124 MBW37445.1 GGFL01013885 MBW78063.1 MWRG01033899 PRD20757.1 JRES01000032 KNC34772.1 KQ971361 EFA05698.1 CP012526 ALC47695.1 GGFK01010643 MBW43964.1 JO841369 AEO32986.1 CH964272 EDW83879.1 KRF99908.1 CH480827 EDW44795.1 ADMH02000540 ETN66036.1 JXUM01148651 KQ570609 KXJ68331.1 CM000364 EDX12625.1 CH940652 EDW58924.2 GDKW01003332 JAI53263.1 UFQT01001180 SSX29357.1 GGMS01005259 MBY74462.1 CH479179 EDW25175.1 CM000070 EDY68158.2 ACPB03011632 LJIJ01000734 ODM94915.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
UP000005204
+ More
UP000037510 UP000001070 UP000007819 UP000002358 UP000215335 UP000009192 UP000037069 UP000007266 UP000092553 UP000007798 UP000192221 UP000001292 UP000000673 UP000069940 UP000249989 UP000000304 UP000008792 UP000069272 UP000008744 UP000001819 UP000015103 UP000094527
UP000037510 UP000001070 UP000007819 UP000002358 UP000215335 UP000009192 UP000037069 UP000007266 UP000092553 UP000007798 UP000192221 UP000001292 UP000000673 UP000069940 UP000249989 UP000000304 UP000008792 UP000069272 UP000008744 UP000001819 UP000015103 UP000094527
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2H1WGR9
A0A2A4IUC0
A0A2W1BYE5
A0A194Q9H2
A0A194Q7Y0
I4DS54
+ More
A0A194RBC8 A0A194QDV6 I4DJ10 A0A3S2NLC3 A0A194RAP6 I4DN32 A0A212ETD1 A0A2A4JI21 A0A2H1W8U7 A0A088MGW5 A0A2W1BT77 A0A2H1W4U4 A0A2H1W9U4 A0A2W1BT67 A0A2A4J7D3 H9JT77 A0A2H1WBG2 H9ISL2 H9JT78 A0A2H1VUX0 A0A194Q3N0 H9ISL1 A0A0N0PED8 H9JQT0 A0A437AS20 A0A437ASV3 A0A3S2L850 A0A3S2NMV9 A0A2H1WAX3 A0A437AYB0 A0A0L7LKV1 A0A2W1BQ93 A0A2A4IUP3 H9IT78 A0A3S2M0V1 A0A2W1BXH1 A0A2W1BS42 A0A2H1UZX5 A0A2A4IUD5 A0A2A4JM77 A0A2A4JMB0 B4JHE0 A0A2L2Y2A3 A0A0A7LYX9 A0A194Q1N0 A0A310SND6 A0A1B6MDD3 J9KYF6 K7IYZ3 A0A232F2X4 B4KC82 A0A2M4CUH8 A0A2M4A9K3 A0A2M4DKI6 A0A2P6K367 A0A0L0CRA6 D2A572 A0A0M4F792 A0A2M4AT71 G3MHL8 B4NJ86 A0A0Q9WVU0 A0A1W4VTT3 B4IBN7 W5JPI5 A0A182G6R1 B4QWD4 B4M5J8 A0A1Y9G9T7 A0A0P4VM15 A0A336MLW0 A0A2S2QB91 B4G4K4 B5DWL8 T1I7W3 A0A1D2MQ77
A0A194RBC8 A0A194QDV6 I4DJ10 A0A3S2NLC3 A0A194RAP6 I4DN32 A0A212ETD1 A0A2A4JI21 A0A2H1W8U7 A0A088MGW5 A0A2W1BT77 A0A2H1W4U4 A0A2H1W9U4 A0A2W1BT67 A0A2A4J7D3 H9JT77 A0A2H1WBG2 H9ISL2 H9JT78 A0A2H1VUX0 A0A194Q3N0 H9ISL1 A0A0N0PED8 H9JQT0 A0A437AS20 A0A437ASV3 A0A3S2L850 A0A3S2NMV9 A0A2H1WAX3 A0A437AYB0 A0A0L7LKV1 A0A2W1BQ93 A0A2A4IUP3 H9IT78 A0A3S2M0V1 A0A2W1BXH1 A0A2W1BS42 A0A2H1UZX5 A0A2A4IUD5 A0A2A4JM77 A0A2A4JMB0 B4JHE0 A0A2L2Y2A3 A0A0A7LYX9 A0A194Q1N0 A0A310SND6 A0A1B6MDD3 J9KYF6 K7IYZ3 A0A232F2X4 B4KC82 A0A2M4CUH8 A0A2M4A9K3 A0A2M4DKI6 A0A2P6K367 A0A0L0CRA6 D2A572 A0A0M4F792 A0A2M4AT71 G3MHL8 B4NJ86 A0A0Q9WVU0 A0A1W4VTT3 B4IBN7 W5JPI5 A0A182G6R1 B4QWD4 B4M5J8 A0A1Y9G9T7 A0A0P4VM15 A0A336MLW0 A0A2S2QB91 B4G4K4 B5DWL8 T1I7W3 A0A1D2MQ77
PDB
6ERC
E-value=3.59395e-06,
Score=118
Ontologies
GO
PANTHER
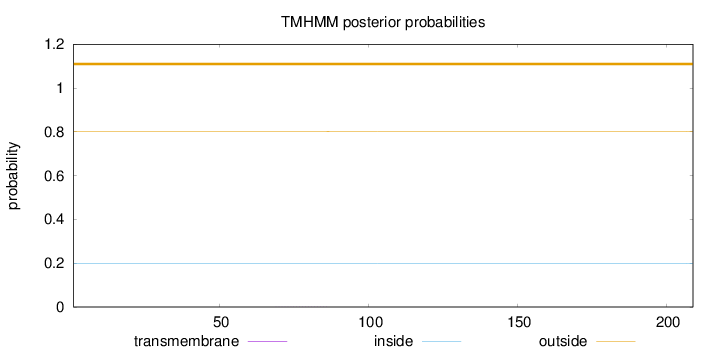
Topology
Length:
209
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00576999999999999
Exp number, first 60 AAs:
0.00115
Total prob of N-in:
0.19775
outside
1 - 209
Population Genetic Test Statistics
Pi
133.440404
Theta
126.688955
Tajima's D
0.131807
CLR
1736.805854
CSRT
0.404379781010949
Interpretation
Uncertain
