Gene
KWMTBOMO12846
Pre Gene Modal
BGIBMGA012754
Annotation
PREDICTED:_palmitoyltransferase_ZDHHC18_[Amyelois_transitella]
Full name
Palmitoyltransferase
+ More
Palmitoyltransferase app
Palmitoyltransferase app
Alternative Name
Protein approximated
Location in the cell
PlasmaMembrane Reliability : 3.874
Sequence
CDS
ATGGCGTCGCGGCGCGTGACGCGCAAGTGGGAAGTGTTCGCGGGGCGAAACCGGTTCTGGTGCGACGGGCGGCTCATGACTGCACCGCACCCCGGCGTCTTCCTGCTCACACTGGCGCTCATCTGCGGCACGTGCGCGCTGCACTTCGCGTTCGACTGTCCCTTCCTGGCCGTGCGCGTGTCGGCCGCCGTGCCCGCCGTGGGCGCCGCGCTGTTCGTGCTGACGCTGGCGGCGCTGCTGCGGACCGCGCTGTCCGACCCGGGCATCATCCCGCGCGCGGCCACGGCCGAGGCGGCGGCCCTGGAGGCTGCCGAAGCGGGTCGGCCTCCGCCGCGCGCGCGGGAGGTGCTCGTGCGAGGCCGCCCCGTCAAGCTCAAGTACTGCTTCACCTGTAAGATGTTCCGACCTCCGCGCGCGTCGCACTGCTCACTCTGCGACAACTGCGTGGACCGGTTCGACCATCACTGTCCGTGGGTGGGGAACTGCGTGGGCAAGCGCAACTACCGGTCGTTCTACACGTTCGTGGTGTCGCTGTCGTTCCTGGCGGTGTTCGTGTTCGCGTGCGCCGTGACGCACCTGGCGCTGGCGGCGCGGGGCGCGGGCGTGGCGGCGGCGCTGCACGCGTCCCCCGCCTCGGCGCTGGTGGCCGCCGTGTGCTTCCTGTCCGTGTGGTCCGTGCTGGGGCTCGCCGGCTTCCACACGTACCTCGCCTCCACCGACCAGACCACCAACGAGGACATTAAAGGTTCATTTTCGAGTCGGCGCGGCGTGTCCAACCCGAACCCGTACTCGCGAGGTAACGCCTGCGCCAACTGCTGGTACGTGCTGTGCGGTCCGCTGGCGCCCAGCCTCATCGACCGGCGCGGCGTGTTCACGATCGACGTGAAGGACGAGCTGCCGCCGAGCTTCGTCCGCGCGCTGGGAGCCGTGACCGTGGCCCCGGCGCCGGGCGGCGGCGCGGAGCCGGCCCGCCACGCCTACATGAACAGGTCGCTGGACGTGGAGCCGGCGGGGGCGGGCGCGGGGGCGGGGGCGGGCGCGGGGGCGGGGGGCGCGGTGGCGCTGAGCGCGTCCCGGCTGCGGCTGCTGCACGACACGACCATGATCGACGCGGCGCTGGACCTGGACGAGCCGGAGCCGCCGCCGCACCTGTCCGTGGCGCTGCCGCCCCTCCCCCCGCACCCGCTCACCGACTGCAACTGA
Protein
MASRRVTRKWEVFAGRNRFWCDGRLMTAPHPGVFLLTLALICGTCALHFAFDCPFLAVRVSAAVPAVGAALFVLTLAALLRTALSDPGIIPRAATAEAAALEAAEAGRPPPRAREVLVRGRPVKLKYCFTCKMFRPPRASHCSLCDNCVDRFDHHCPWVGNCVGKRNYRSFYTFVVSLSFLAVFVFACAVTHLALAARGAGVAAALHASPASALVAAVCFLSVWSVLGLAGFHTYLASTDQTTNEDIKGSFSSRRGVSNPNPYSRGNACANCWYVLCGPLAPSLIDRRGVFTIDVKDELPPSFVRALGAVTVAPAPGGGAEPARHAYMNRSLDVEPAGAGAGAGAGAGAGGAVALSASRLRLLHDTTMIDAALDLDEPEPPPHLSVALPPLPPHPLTDCN
Summary
Description
Palmitoylates Dlish which is required for the apical cell cortex localization, total cellular level and full activity of dachs.
Catalytic Activity
hexadecanoyl-CoA + L-cysteinyl-[protein] = CoA + S-hexadecanoyl-L-cysteinyl-[protein]
Subunit
Interacts with Dlish (via N-terminus including SH3 domain 1); this leads to palmitoylation of Dlish by app. Interacts with ft; this is likely to interfere with the interaction between app and Dlish, reducing the ability of app to palmitoylate Dlish and tether it to the apical cell cortex.
Similarity
Belongs to the DHHC palmitoyltransferase family.
Belongs to the DHHC palmitoyltransferase family. ERF2/ZDHHC9 subfamily.
Belongs to the actin family.
Belongs to the DHHC palmitoyltransferase family. ERF2/ZDHHC9 subfamily.
Belongs to the actin family.
Keywords
Acyltransferase
Alternative splicing
Cell membrane
Complete proteome
Endoplasmic reticulum
Lipoprotein
Membrane
Palmitate
Reference proteome
Transferase
Transmembrane
Transmembrane helix
Feature
chain Palmitoyltransferase app
splice variant In isoform S.
splice variant In isoform S.
Uniprot
A0A2H1V743
A0A3S2NK73
A0A212FI60
D6WMN7
A0A0T6B0T6
T1DPQ6
+ More
A0A0N8P0N9 A0A182S9E3 A0A1I8MH71 A0A1I8MH67 A0A0R3P3F1 A0A0R1DXL5 A0A0Q5U6K8 A2VEY9-4 A2VEY9-3 A0A0J9UJS1 Q6NLJ6 A0A182GGI1 A0A0K8VB96 A0A1Q3G1M0 A0A1Y1L9P1 A0A1Q3G1S1 A0A2M3Z709 A0A1J1IZW7 A0A1Q3G1K9 A0A1Q3G1V4 A0A1Q3G1S9 A0A1Q3G1R9 A0A1Q3G1L2 A0A0J9RV84 A0A1Q3G1H7 A0A1Q3G1N8 A0A1Q3G1E5 A0A2M4AKJ3 T1E917 A0A2M3Z4A0 A0A1Y1L793 A0A0A1XP17 A0A0A1WSW5 A0A034WAW4 A0A0P8XSI6 A0A0L7QT25 W8ANY2 B4MLG2 A0A067QNT6 A0A0Q9WI55 A0A1W4UV67 A0A1I8QB68 A0A1I8MH70 A0A3B0J8S2 A0A1S4EYN0 A0A0R1E2F2 Q17LD8 A0A0Q5UAV8 B4J344 A0A2M4CTD3 A0A023EUZ5 A0A0A1XGZ5 A0A0J9RTU2 Q2M0R3 B3M7U3 A2VEY9 A0A2M4BEX1 A0A0A1WQD9 E0W385 A0A0K8V2I3 A0A0K8TYX9 A0A2M4BE79 B4LFF0 A0A1W4UTH4 B4QR78 A0A0J9RU84 Q9VTV6 B4KZZ3 A0A3B0J7V9 B3NH86 B4PGB3 V9I969 E2BKR2 A0A3L8DFQ4 A0A026W9J6 A0A084W9L5 D0QWS3 A0A453YJN4 F5HLQ3 A0A1B6CPV3 A0A0L0BUZ1 A0A151WSA0 F4X279 B4HFK6 A0A336KRX4 E2A947 A0A151INJ4 A0A182YFL5 V9IAI1 A0A182FCF7 A0A182J3X3 A0A2A3EF37 W5JTX3 A0A182QCH5
A0A0N8P0N9 A0A182S9E3 A0A1I8MH71 A0A1I8MH67 A0A0R3P3F1 A0A0R1DXL5 A0A0Q5U6K8 A2VEY9-4 A2VEY9-3 A0A0J9UJS1 Q6NLJ6 A0A182GGI1 A0A0K8VB96 A0A1Q3G1M0 A0A1Y1L9P1 A0A1Q3G1S1 A0A2M3Z709 A0A1J1IZW7 A0A1Q3G1K9 A0A1Q3G1V4 A0A1Q3G1S9 A0A1Q3G1R9 A0A1Q3G1L2 A0A0J9RV84 A0A1Q3G1H7 A0A1Q3G1N8 A0A1Q3G1E5 A0A2M4AKJ3 T1E917 A0A2M3Z4A0 A0A1Y1L793 A0A0A1XP17 A0A0A1WSW5 A0A034WAW4 A0A0P8XSI6 A0A0L7QT25 W8ANY2 B4MLG2 A0A067QNT6 A0A0Q9WI55 A0A1W4UV67 A0A1I8QB68 A0A1I8MH70 A0A3B0J8S2 A0A1S4EYN0 A0A0R1E2F2 Q17LD8 A0A0Q5UAV8 B4J344 A0A2M4CTD3 A0A023EUZ5 A0A0A1XGZ5 A0A0J9RTU2 Q2M0R3 B3M7U3 A2VEY9 A0A2M4BEX1 A0A0A1WQD9 E0W385 A0A0K8V2I3 A0A0K8TYX9 A0A2M4BE79 B4LFF0 A0A1W4UTH4 B4QR78 A0A0J9RU84 Q9VTV6 B4KZZ3 A0A3B0J7V9 B3NH86 B4PGB3 V9I969 E2BKR2 A0A3L8DFQ4 A0A026W9J6 A0A084W9L5 D0QWS3 A0A453YJN4 F5HLQ3 A0A1B6CPV3 A0A0L0BUZ1 A0A151WSA0 F4X279 B4HFK6 A0A336KRX4 E2A947 A0A151INJ4 A0A182YFL5 V9IAI1 A0A182FCF7 A0A182J3X3 A0A2A3EF37 W5JTX3 A0A182QCH5
EC Number
2.3.1.225
Pubmed
22118469
18362917
19820115
17994087
25315136
15632085
+ More
17550304 10731132 12537572 18804377 18719403 27692068 27824307 22936249 26483478 28004739 25830018 25348373 24495485 24845553 17510324 18057021 24945155 23185243 20566863 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 30249741 24508170 24438588 19859648 12364791 14747013 17210077 26108605 21719571 25244985 20920257 23761445
17550304 10731132 12537572 18804377 18719403 27692068 27824307 22936249 26483478 28004739 25830018 25348373 24495485 24845553 17510324 18057021 24945155 23185243 20566863 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20798317 30249741 24508170 24438588 19859648 12364791 14747013 17210077 26108605 21719571 25244985 20920257 23761445
EMBL
ODYU01000773
SOQ36074.1
RSAL01000015
RVE53071.1
AGBW02008428
OWR53424.1
+ More
KQ971343 EFA04303.2 LJIG01016404 KRT80702.1 GAMD01001730 JAA99860.1 CH902618 KPU77652.1 CH379069 KRT07714.1 CM000159 KRK01733.1 CH954178 KQS43836.1 AE014296 BT030308 ABN49447.1 CM002912 KMY99200.1 BT012335 AAS77460.1 JXUM01011106 JXUM01011107 JXUM01011108 JXUM01011109 JXUM01011110 JXUM01011111 JXUM01011112 JXUM01011113 JXUM01011114 JXUM01011115 GDHF01026100 GDHF01017497 GDHF01016168 JAI26214.1 JAI34817.1 JAI36146.1 GFDL01001375 JAV33670.1 GEZM01062665 JAV69551.1 GFDL01001356 JAV33689.1 GGFM01003556 MBW24307.1 CVRI01000063 CRL04718.1 GFDL01001353 JAV33692.1 GFDL01001316 JAV33729.1 GFDL01001346 JAV33699.1 GFDL01001339 JAV33706.1 GFDL01001374 JAV33671.1 KMY99199.1 GFDL01001387 JAV33658.1 GFDL01001359 JAV33686.1 GFDL01001421 JAV33624.1 GGFK01007931 MBW41252.1 GAMD01001727 JAA99863.1 GGFM01002600 MBW23351.1 GEZM01062663 JAV69553.1 GBXI01001662 JAD12630.1 GBXI01012779 JAD01513.1 GAKP01008005 GAKP01008004 GAKP01008003 GAKP01008002 JAC50948.1 KPU77654.1 KQ414755 KOC61792.1 GAMC01016090 JAB90465.1 CH963847 EDW72818.2 KK853114 KDR11201.1 CH940647 KRF84264.1 OUUW01000002 SPP76282.1 KRK01732.1 CH477216 EAT47524.1 KQS43837.1 CH916366 EDV96115.1 GGFL01003910 MBW68088.1 GAPW01000328 JAC13270.1 GBXI01017124 GBXI01004065 JAC97167.1 JAD10227.1 KMY99201.1 EAL30867.3 EDV38816.2 GGFJ01002227 MBW51368.1 GBXI01013028 JAD01264.1 DS235882 EEB20091.1 GDHF01019258 GDHF01005678 JAI33056.1 JAI46636.1 GDHF01032828 GDHF01024048 JAI19486.1 JAI28266.1 GGFJ01002218 MBW51359.1 EDW69248.1 KRF84263.1 CM000363 EDX10208.1 KMY99197.1 AAF49939.3 CH933809 EDW18005.2 SPP76283.1 EDV51543.2 EDW94277.2 JR037217 AEY57643.1 GL448819 EFN83760.1 QOIP01000009 RLU19022.1 KK107320 EZA52782.1 ATLV01021831 KE525324 KFB46909.1 FJ821035 ACN94781.1 APCN01002922 APCN01002923 AAAB01008900 EGK97214.1 GEDC01021894 JAS15404.1 JRES01001294 KNC23857.1 KQ982779 KYQ50736.1 GL888565 EGI59495.1 CH480815 EDW41237.1 UFQS01000784 UFQT01000784 SSX06868.1 SSX27212.1 GL437711 EFN70032.1 KQ976932 KYN07042.1 JR037216 AEY57642.1 KZ288263 PBC30328.1 ADMH02000391 ETN66738.1 AXCN02000912
KQ971343 EFA04303.2 LJIG01016404 KRT80702.1 GAMD01001730 JAA99860.1 CH902618 KPU77652.1 CH379069 KRT07714.1 CM000159 KRK01733.1 CH954178 KQS43836.1 AE014296 BT030308 ABN49447.1 CM002912 KMY99200.1 BT012335 AAS77460.1 JXUM01011106 JXUM01011107 JXUM01011108 JXUM01011109 JXUM01011110 JXUM01011111 JXUM01011112 JXUM01011113 JXUM01011114 JXUM01011115 GDHF01026100 GDHF01017497 GDHF01016168 JAI26214.1 JAI34817.1 JAI36146.1 GFDL01001375 JAV33670.1 GEZM01062665 JAV69551.1 GFDL01001356 JAV33689.1 GGFM01003556 MBW24307.1 CVRI01000063 CRL04718.1 GFDL01001353 JAV33692.1 GFDL01001316 JAV33729.1 GFDL01001346 JAV33699.1 GFDL01001339 JAV33706.1 GFDL01001374 JAV33671.1 KMY99199.1 GFDL01001387 JAV33658.1 GFDL01001359 JAV33686.1 GFDL01001421 JAV33624.1 GGFK01007931 MBW41252.1 GAMD01001727 JAA99863.1 GGFM01002600 MBW23351.1 GEZM01062663 JAV69553.1 GBXI01001662 JAD12630.1 GBXI01012779 JAD01513.1 GAKP01008005 GAKP01008004 GAKP01008003 GAKP01008002 JAC50948.1 KPU77654.1 KQ414755 KOC61792.1 GAMC01016090 JAB90465.1 CH963847 EDW72818.2 KK853114 KDR11201.1 CH940647 KRF84264.1 OUUW01000002 SPP76282.1 KRK01732.1 CH477216 EAT47524.1 KQS43837.1 CH916366 EDV96115.1 GGFL01003910 MBW68088.1 GAPW01000328 JAC13270.1 GBXI01017124 GBXI01004065 JAC97167.1 JAD10227.1 KMY99201.1 EAL30867.3 EDV38816.2 GGFJ01002227 MBW51368.1 GBXI01013028 JAD01264.1 DS235882 EEB20091.1 GDHF01019258 GDHF01005678 JAI33056.1 JAI46636.1 GDHF01032828 GDHF01024048 JAI19486.1 JAI28266.1 GGFJ01002218 MBW51359.1 EDW69248.1 KRF84263.1 CM000363 EDX10208.1 KMY99197.1 AAF49939.3 CH933809 EDW18005.2 SPP76283.1 EDV51543.2 EDW94277.2 JR037217 AEY57643.1 GL448819 EFN83760.1 QOIP01000009 RLU19022.1 KK107320 EZA52782.1 ATLV01021831 KE525324 KFB46909.1 FJ821035 ACN94781.1 APCN01002922 APCN01002923 AAAB01008900 EGK97214.1 GEDC01021894 JAS15404.1 JRES01001294 KNC23857.1 KQ982779 KYQ50736.1 GL888565 EGI59495.1 CH480815 EDW41237.1 UFQS01000784 UFQT01000784 SSX06868.1 SSX27212.1 GL437711 EFN70032.1 KQ976932 KYN07042.1 JR037216 AEY57642.1 KZ288263 PBC30328.1 ADMH02000391 ETN66738.1 AXCN02000912
Proteomes
UP000283053
UP000007151
UP000007266
UP000007801
UP000075901
UP000095301
+ More
UP000001819 UP000002282 UP000008711 UP000000803 UP000069940 UP000183832 UP000053825 UP000007798 UP000027135 UP000008792 UP000192221 UP000095300 UP000268350 UP000008820 UP000001070 UP000009046 UP000000304 UP000009192 UP000008237 UP000279307 UP000053097 UP000030765 UP000075840 UP000007062 UP000037069 UP000075809 UP000007755 UP000001292 UP000000311 UP000078542 UP000076408 UP000069272 UP000075880 UP000242457 UP000000673 UP000075886
UP000001819 UP000002282 UP000008711 UP000000803 UP000069940 UP000183832 UP000053825 UP000007798 UP000027135 UP000008792 UP000192221 UP000095300 UP000268350 UP000008820 UP000001070 UP000009046 UP000000304 UP000009192 UP000008237 UP000279307 UP000053097 UP000030765 UP000075840 UP000007062 UP000037069 UP000075809 UP000007755 UP000001292 UP000000311 UP000078542 UP000076408 UP000069272 UP000075880 UP000242457 UP000000673 UP000075886
Interpro
ProteinModelPortal
A0A2H1V743
A0A3S2NK73
A0A212FI60
D6WMN7
A0A0T6B0T6
T1DPQ6
+ More
A0A0N8P0N9 A0A182S9E3 A0A1I8MH71 A0A1I8MH67 A0A0R3P3F1 A0A0R1DXL5 A0A0Q5U6K8 A2VEY9-4 A2VEY9-3 A0A0J9UJS1 Q6NLJ6 A0A182GGI1 A0A0K8VB96 A0A1Q3G1M0 A0A1Y1L9P1 A0A1Q3G1S1 A0A2M3Z709 A0A1J1IZW7 A0A1Q3G1K9 A0A1Q3G1V4 A0A1Q3G1S9 A0A1Q3G1R9 A0A1Q3G1L2 A0A0J9RV84 A0A1Q3G1H7 A0A1Q3G1N8 A0A1Q3G1E5 A0A2M4AKJ3 T1E917 A0A2M3Z4A0 A0A1Y1L793 A0A0A1XP17 A0A0A1WSW5 A0A034WAW4 A0A0P8XSI6 A0A0L7QT25 W8ANY2 B4MLG2 A0A067QNT6 A0A0Q9WI55 A0A1W4UV67 A0A1I8QB68 A0A1I8MH70 A0A3B0J8S2 A0A1S4EYN0 A0A0R1E2F2 Q17LD8 A0A0Q5UAV8 B4J344 A0A2M4CTD3 A0A023EUZ5 A0A0A1XGZ5 A0A0J9RTU2 Q2M0R3 B3M7U3 A2VEY9 A0A2M4BEX1 A0A0A1WQD9 E0W385 A0A0K8V2I3 A0A0K8TYX9 A0A2M4BE79 B4LFF0 A0A1W4UTH4 B4QR78 A0A0J9RU84 Q9VTV6 B4KZZ3 A0A3B0J7V9 B3NH86 B4PGB3 V9I969 E2BKR2 A0A3L8DFQ4 A0A026W9J6 A0A084W9L5 D0QWS3 A0A453YJN4 F5HLQ3 A0A1B6CPV3 A0A0L0BUZ1 A0A151WSA0 F4X279 B4HFK6 A0A336KRX4 E2A947 A0A151INJ4 A0A182YFL5 V9IAI1 A0A182FCF7 A0A182J3X3 A0A2A3EF37 W5JTX3 A0A182QCH5
A0A0N8P0N9 A0A182S9E3 A0A1I8MH71 A0A1I8MH67 A0A0R3P3F1 A0A0R1DXL5 A0A0Q5U6K8 A2VEY9-4 A2VEY9-3 A0A0J9UJS1 Q6NLJ6 A0A182GGI1 A0A0K8VB96 A0A1Q3G1M0 A0A1Y1L9P1 A0A1Q3G1S1 A0A2M3Z709 A0A1J1IZW7 A0A1Q3G1K9 A0A1Q3G1V4 A0A1Q3G1S9 A0A1Q3G1R9 A0A1Q3G1L2 A0A0J9RV84 A0A1Q3G1H7 A0A1Q3G1N8 A0A1Q3G1E5 A0A2M4AKJ3 T1E917 A0A2M3Z4A0 A0A1Y1L793 A0A0A1XP17 A0A0A1WSW5 A0A034WAW4 A0A0P8XSI6 A0A0L7QT25 W8ANY2 B4MLG2 A0A067QNT6 A0A0Q9WI55 A0A1W4UV67 A0A1I8QB68 A0A1I8MH70 A0A3B0J8S2 A0A1S4EYN0 A0A0R1E2F2 Q17LD8 A0A0Q5UAV8 B4J344 A0A2M4CTD3 A0A023EUZ5 A0A0A1XGZ5 A0A0J9RTU2 Q2M0R3 B3M7U3 A2VEY9 A0A2M4BEX1 A0A0A1WQD9 E0W385 A0A0K8V2I3 A0A0K8TYX9 A0A2M4BE79 B4LFF0 A0A1W4UTH4 B4QR78 A0A0J9RU84 Q9VTV6 B4KZZ3 A0A3B0J7V9 B3NH86 B4PGB3 V9I969 E2BKR2 A0A3L8DFQ4 A0A026W9J6 A0A084W9L5 D0QWS3 A0A453YJN4 F5HLQ3 A0A1B6CPV3 A0A0L0BUZ1 A0A151WSA0 F4X279 B4HFK6 A0A336KRX4 E2A947 A0A151INJ4 A0A182YFL5 V9IAI1 A0A182FCF7 A0A182J3X3 A0A2A3EF37 W5JTX3 A0A182QCH5
PDB
6BML
E-value=1.41828e-12,
Score=176
Ontologies
GO
PANTHER
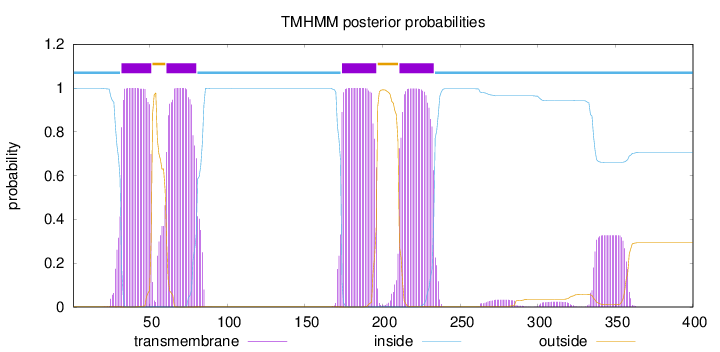
Topology
Subcellular location
Endoplasmic reticulum membrane
Apical cell membrane
Apical cell membrane
Length:
400
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
96.64138
Exp number, first 60 AAs:
22.81608
Total prob of N-in:
0.99883
POSSIBLE N-term signal
sequence
inside
1 - 31
TMhelix
32 - 51
outside
52 - 60
TMhelix
61 - 80
inside
81 - 173
TMhelix
174 - 196
outside
197 - 210
TMhelix
211 - 233
inside
234 - 400
Population Genetic Test Statistics
Pi
219.556349
Theta
185.014064
Tajima's D
0.688159
CLR
0.002844
CSRT
0.569821508924554
Interpretation
Uncertain
