Gene
KWMTBOMO12845
Pre Gene Modal
BGIBMGA012718
Annotation
triose-phosphate_transporter-like_protein_[Bombyx_mori]
Full name
Solute carrier family 35 member E1 homolog
Location in the cell
PlasmaMembrane Reliability : 3.48
Sequence
CDS
ATGGGTACAAGCGGCTCCAGGCGCGAGACACTAATAGTTGGCTTCCTGTGCGCAGCGTGGTATATGTTAAGTTCAGCCAGTAATGTCGTGGGCAAACTGGCATTAACAGAGCTACCATTTCCACTAACTATGACAGCGGTACAGCTGTGTGCAGCGGCCTCGCTGAGTGTACCCGCGCTAGCCCTGTGTGGAGTACGTTCGACGCGATGGCCGACGAACTATTGGACCCGTGTCTTAGTACCCTTGGCGATCGCCAAACTTCTTACCACACTGTGCTCGCAAGTTTCCATTTGGAAAGTGCCAGTCTCCTATGCCCACACAGTGAAAGCGACGACTCCTCTCTGGACGGCGGGGCTGGCTCGCGTACTGTTTGGGGAGCGCGTGTCTCGCGGCGTGGCGGGCGCGCTGCTCGTGATCGCGGGCGGGGTCGCGCTCGCCTCGCTCACCGAGCTGCAGTTCGATGCGCTGGGGCTGGGAGCCGCGCTCACGTCCGCCGCGCTGTTGGCGCTGCAGCATCTGTATTCGAAGCGCGCGCTGCAGGACTCCGGCGTGCACCACTTGCGACTGCTGGCGACGTTGTCGGGTCTGGCGCTGGTGCCGATGGCTCCGTTGTGGTTGGTGCGGGACGCCGGCGCCGTACTGCGCGCCCAGGTGGCGTGGAACCGCGCGGGCCCGCTGCTGCTGGCGGACGGGGTGCTGGCGTGGCTGCAGGCGGTGGCTGCGTTCTCGGTGTTGTCGCGAGTGTCGCCGCTGACGTACTCAGTGGCGTCGGCTGCGAAGCGCGCCGTGGTGGTGGGCGCGTCGCTGGTGGTGCTGAGGAACCCGGCGCCGCCGCTGAACGTGGTGGGCATGTCGGTGGCAGTGCTGGGTGTGCTGGCTTATGACCGAGCCCGAGCGGCCGCCCGCCGCGCCCCCCGCCCCGCGCTGCCAGTCTGA
Protein
MGTSGSRRETLIVGFLCAAWYMLSSASNVVGKLALTELPFPLTMTAVQLCAAASLSVPALALCGVRSTRWPTNYWTRVLVPLAIAKLLTTLCSQVSIWKVPVSYAHTVKATTPLWTAGLARVLFGERVSRGVAGALLVIAGGVALASLTELQFDALGLGAALTSAALLALQHLYSKRALQDSGVHHLRLLATLSGLALVPMAPLWLVRDAGAVLRAQVAWNRAGPLLLADGVLAWLQAVAAFSVLSRVSPLTYSVASAAKRAVVVGASLVVLRNPAPPLNVVGMSVAVLGVLAYDRARAAARRAPRPALPV
Summary
Description
Putative transporter.
Similarity
Belongs to the TPT transporter family. SLC35E subfamily.
Keywords
Complete proteome
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Solute carrier family 35 member E1 homolog
Uniprot
D7R261
A0A194Q9J4
A0A3S2NZG6
A0A212FI58
A0A154PRP0
A0A0M9A9Z6
+ More
A0A087ZSM6 A0A2A3EEM6 A0A2P8Y3U1 E2BCI5 U4UQ50 N6UMK4 B0W9K6 A0A1Y1MH74 A0A0T6B3K2 B4L2V1 A0A1Q3F8C4 A0A1Q3F8A1 A0A1Q3F8G2 A0A195C287 A0A182JIP4 A0A182N1A8 A0A182JSU1 A0A3L8E179 B4JWR7 A0A182W4N4 U5ENR4 A0A182SMI5 A0A182YMY7 A0A084VX09 B4M379 A0A182RBI4 A0A151IR96 A0A1B6ISH0 A0A026WY37 A0A1B6KR95 A0A182XLT3 F4WUF7 A0A182IA91 A0A1S4G957 A0A158NN40 A0A1B6EKM7 B4ILC9 X2JLL7 Q9VR50 A0A182USS5 A0A151X0C3 A0A1A9X8T0 B3NYG8 B4N1K1 A0A1I8MKA0 Q7QFK8 A0A0M4EJT5 B4PZ64 Q29JF1 B4H301 A0A151HYP1 A0A182Q475 A0A182TUW8 A0A1A9Z2E3 A0A1A9VRL1 A0A023ER00 A0A1W4VWR4 B3MRT5 A0A3B0J6N3 A0A182HEA2 A0A1A9WCB4 A0A151JVG2 A0A182P7W1 A0A0K8VZ88 A0A224XRT3 A0A1I8QB48 A0A1J1HQF7 A0A1B0FAE1 A0A0A1WFX9 W5JDZ2 A0A034VKC8 A0A3R7PW09 A0A2M3Z542 A0A182FJ28 A0A0P4VWQ0 T1HZ76 A0A067RDZ0 A0A1B0CHN3 A0A0L0C7Z7 A0A2R7VWL4 A0A1L8DDU9 A0A0P6HIW7 A0A0P5HU50 D6X3C6 A0A2M4BME2 A0A2M4BMD4 A0A0A9XPU8 A0A2M4BN94 E2A8A7 A0A0L7QT22 A0A146KTF9 A0A0K8WAK3 A0A0P5NMU8
A0A087ZSM6 A0A2A3EEM6 A0A2P8Y3U1 E2BCI5 U4UQ50 N6UMK4 B0W9K6 A0A1Y1MH74 A0A0T6B3K2 B4L2V1 A0A1Q3F8C4 A0A1Q3F8A1 A0A1Q3F8G2 A0A195C287 A0A182JIP4 A0A182N1A8 A0A182JSU1 A0A3L8E179 B4JWR7 A0A182W4N4 U5ENR4 A0A182SMI5 A0A182YMY7 A0A084VX09 B4M379 A0A182RBI4 A0A151IR96 A0A1B6ISH0 A0A026WY37 A0A1B6KR95 A0A182XLT3 F4WUF7 A0A182IA91 A0A1S4G957 A0A158NN40 A0A1B6EKM7 B4ILC9 X2JLL7 Q9VR50 A0A182USS5 A0A151X0C3 A0A1A9X8T0 B3NYG8 B4N1K1 A0A1I8MKA0 Q7QFK8 A0A0M4EJT5 B4PZ64 Q29JF1 B4H301 A0A151HYP1 A0A182Q475 A0A182TUW8 A0A1A9Z2E3 A0A1A9VRL1 A0A023ER00 A0A1W4VWR4 B3MRT5 A0A3B0J6N3 A0A182HEA2 A0A1A9WCB4 A0A151JVG2 A0A182P7W1 A0A0K8VZ88 A0A224XRT3 A0A1I8QB48 A0A1J1HQF7 A0A1B0FAE1 A0A0A1WFX9 W5JDZ2 A0A034VKC8 A0A3R7PW09 A0A2M3Z542 A0A182FJ28 A0A0P4VWQ0 T1HZ76 A0A067RDZ0 A0A1B0CHN3 A0A0L0C7Z7 A0A2R7VWL4 A0A1L8DDU9 A0A0P6HIW7 A0A0P5HU50 D6X3C6 A0A2M4BME2 A0A2M4BMD4 A0A0A9XPU8 A0A2M4BN94 E2A8A7 A0A0L7QT22 A0A146KTF9 A0A0K8WAK3 A0A0P5NMU8
Pubmed
19121390
26354079
22118469
29403074
20798317
23537049
+ More
28004739 17994087 30249741 25244985 24438588 24508170 21719571 12364791 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304 15632085 24945155 26483478 25830018 20920257 23761445 25348373 27129103 24845553 26108605 18362917 19820115 25401762 26823975
28004739 17994087 30249741 25244985 24438588 24508170 21719571 12364791 21347285 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25315136 17550304 15632085 24945155 26483478 25830018 20920257 23761445 25348373 27129103 24845553 26108605 18362917 19820115 25401762 26823975
EMBL
BABH01034908
HM026603
ADG85767.1
KQ459324
KPJ01680.1
RSAL01000015
+ More
RVE53073.1 AGBW02008428 OWR53423.1 KQ435090 KZC14561.1 KQ435718 KOX78983.1 KZ288266 PBC30187.1 PYGN01000958 PSN38915.1 GL447283 EFN86611.1 KB632318 ERL92266.1 APGK01017166 APGK01017167 KB739998 ENN81931.1 DS231864 EDS40231.1 GEZM01031310 JAV84941.1 LJIG01016018 KRT81860.1 CH933810 EDW06918.2 GFDL01011249 JAV23796.1 GFDL01011250 JAV23795.1 GFDL01011203 JAV23842.1 KQ978350 KYM94700.1 QOIP01000002 RLU26145.1 CH916376 EDV95193.1 GANO01000483 JAB59388.1 ATLV01017829 KE525195 KFB42503.1 CH940651 EDW65254.1 KQ981148 KYN09089.1 GECU01017838 JAS89868.1 KK107069 EZA60738.1 GEBQ01026020 JAT13957.1 GL888363 EGI62162.1 APCN01001132 AAAB01008846 ADTU01002501 GECZ01031337 JAS38432.1 CH480870 EDW53788.1 AE014298 AHN59992.1 AHN59993.1 BT003539 KQ982617 KYQ53806.1 CH954180 EDV47647.1 CH963925 EDW78240.1 EAA06186.4 CP012528 ALC49466.1 CM000162 EDX03125.1 KRK07128.1 CH379063 EAL32350.2 CH479205 EDW30833.1 KQ976715 KYM76908.1 AXCN02001240 GAPW01002045 JAC11553.1 CH902622 EDV34490.1 OUUW01000003 SPP77834.1 JXUM01131221 KQ567682 KXJ69321.1 KQ981695 KYN37540.1 GDHF01017416 GDHF01008419 JAI34898.1 JAI43895.1 GFTR01005276 JAW11150.1 CVRI01000014 CRK89762.1 CCAG010015200 GBXI01016902 JAC97389.1 ADMH02001819 ETN61019.1 GAKP01016939 JAC42013.1 QCYY01001330 ROT78803.1 GGFM01002872 MBW23623.1 GDKW01002575 JAI54020.1 ACPB03021330 KK852523 KDR22076.1 AJWK01012538 JRES01000781 KNC28365.1 KK854133 PTY11852.1 GFDF01009537 JAV04547.1 GDIQ01027368 JAN67369.1 GDIQ01228062 GDIP01090063 JAK23663.1 JAM13652.1 KQ971372 EFA09770.1 GGFJ01005109 MBW54250.1 GGFJ01005108 MBW54249.1 GBHO01021605 GBRD01012632 JAG21999.1 JAG53192.1 GGFJ01005107 MBW54248.1 GL437526 EFN70320.1 KQ414756 KOC61621.1 GDHC01020162 JAP98466.1 GDHF01004454 JAI47860.1 GDIQ01142845 JAL08881.1
RVE53073.1 AGBW02008428 OWR53423.1 KQ435090 KZC14561.1 KQ435718 KOX78983.1 KZ288266 PBC30187.1 PYGN01000958 PSN38915.1 GL447283 EFN86611.1 KB632318 ERL92266.1 APGK01017166 APGK01017167 KB739998 ENN81931.1 DS231864 EDS40231.1 GEZM01031310 JAV84941.1 LJIG01016018 KRT81860.1 CH933810 EDW06918.2 GFDL01011249 JAV23796.1 GFDL01011250 JAV23795.1 GFDL01011203 JAV23842.1 KQ978350 KYM94700.1 QOIP01000002 RLU26145.1 CH916376 EDV95193.1 GANO01000483 JAB59388.1 ATLV01017829 KE525195 KFB42503.1 CH940651 EDW65254.1 KQ981148 KYN09089.1 GECU01017838 JAS89868.1 KK107069 EZA60738.1 GEBQ01026020 JAT13957.1 GL888363 EGI62162.1 APCN01001132 AAAB01008846 ADTU01002501 GECZ01031337 JAS38432.1 CH480870 EDW53788.1 AE014298 AHN59992.1 AHN59993.1 BT003539 KQ982617 KYQ53806.1 CH954180 EDV47647.1 CH963925 EDW78240.1 EAA06186.4 CP012528 ALC49466.1 CM000162 EDX03125.1 KRK07128.1 CH379063 EAL32350.2 CH479205 EDW30833.1 KQ976715 KYM76908.1 AXCN02001240 GAPW01002045 JAC11553.1 CH902622 EDV34490.1 OUUW01000003 SPP77834.1 JXUM01131221 KQ567682 KXJ69321.1 KQ981695 KYN37540.1 GDHF01017416 GDHF01008419 JAI34898.1 JAI43895.1 GFTR01005276 JAW11150.1 CVRI01000014 CRK89762.1 CCAG010015200 GBXI01016902 JAC97389.1 ADMH02001819 ETN61019.1 GAKP01016939 JAC42013.1 QCYY01001330 ROT78803.1 GGFM01002872 MBW23623.1 GDKW01002575 JAI54020.1 ACPB03021330 KK852523 KDR22076.1 AJWK01012538 JRES01000781 KNC28365.1 KK854133 PTY11852.1 GFDF01009537 JAV04547.1 GDIQ01027368 JAN67369.1 GDIQ01228062 GDIP01090063 JAK23663.1 JAM13652.1 KQ971372 EFA09770.1 GGFJ01005109 MBW54250.1 GGFJ01005108 MBW54249.1 GBHO01021605 GBRD01012632 JAG21999.1 JAG53192.1 GGFJ01005107 MBW54248.1 GL437526 EFN70320.1 KQ414756 KOC61621.1 GDHC01020162 JAP98466.1 GDHF01004454 JAI47860.1 GDIQ01142845 JAL08881.1
Proteomes
UP000005204
UP000053268
UP000283053
UP000007151
UP000076502
UP000053105
+ More
UP000005203 UP000242457 UP000245037 UP000008237 UP000030742 UP000019118 UP000002320 UP000009192 UP000078542 UP000075880 UP000075884 UP000075881 UP000279307 UP000001070 UP000075920 UP000075901 UP000076408 UP000030765 UP000008792 UP000075900 UP000078492 UP000053097 UP000076407 UP000007755 UP000075840 UP000005205 UP000001292 UP000000803 UP000075903 UP000075809 UP000092443 UP000008711 UP000007798 UP000095301 UP000007062 UP000092553 UP000002282 UP000001819 UP000008744 UP000078540 UP000075886 UP000075902 UP000092445 UP000078200 UP000192221 UP000007801 UP000268350 UP000069940 UP000249989 UP000091820 UP000078541 UP000075885 UP000095300 UP000183832 UP000092444 UP000000673 UP000283509 UP000069272 UP000015103 UP000027135 UP000092461 UP000037069 UP000007266 UP000000311 UP000053825
UP000005203 UP000242457 UP000245037 UP000008237 UP000030742 UP000019118 UP000002320 UP000009192 UP000078542 UP000075880 UP000075884 UP000075881 UP000279307 UP000001070 UP000075920 UP000075901 UP000076408 UP000030765 UP000008792 UP000075900 UP000078492 UP000053097 UP000076407 UP000007755 UP000075840 UP000005205 UP000001292 UP000000803 UP000075903 UP000075809 UP000092443 UP000008711 UP000007798 UP000095301 UP000007062 UP000092553 UP000002282 UP000001819 UP000008744 UP000078540 UP000075886 UP000075902 UP000092445 UP000078200 UP000192221 UP000007801 UP000268350 UP000069940 UP000249989 UP000091820 UP000078541 UP000075885 UP000095300 UP000183832 UP000092444 UP000000673 UP000283509 UP000069272 UP000015103 UP000027135 UP000092461 UP000037069 UP000007266 UP000000311 UP000053825
Pfam
PF03151 TPT
Interpro
IPR004853
Sugar_P_trans_dom
ProteinModelPortal
D7R261
A0A194Q9J4
A0A3S2NZG6
A0A212FI58
A0A154PRP0
A0A0M9A9Z6
+ More
A0A087ZSM6 A0A2A3EEM6 A0A2P8Y3U1 E2BCI5 U4UQ50 N6UMK4 B0W9K6 A0A1Y1MH74 A0A0T6B3K2 B4L2V1 A0A1Q3F8C4 A0A1Q3F8A1 A0A1Q3F8G2 A0A195C287 A0A182JIP4 A0A182N1A8 A0A182JSU1 A0A3L8E179 B4JWR7 A0A182W4N4 U5ENR4 A0A182SMI5 A0A182YMY7 A0A084VX09 B4M379 A0A182RBI4 A0A151IR96 A0A1B6ISH0 A0A026WY37 A0A1B6KR95 A0A182XLT3 F4WUF7 A0A182IA91 A0A1S4G957 A0A158NN40 A0A1B6EKM7 B4ILC9 X2JLL7 Q9VR50 A0A182USS5 A0A151X0C3 A0A1A9X8T0 B3NYG8 B4N1K1 A0A1I8MKA0 Q7QFK8 A0A0M4EJT5 B4PZ64 Q29JF1 B4H301 A0A151HYP1 A0A182Q475 A0A182TUW8 A0A1A9Z2E3 A0A1A9VRL1 A0A023ER00 A0A1W4VWR4 B3MRT5 A0A3B0J6N3 A0A182HEA2 A0A1A9WCB4 A0A151JVG2 A0A182P7W1 A0A0K8VZ88 A0A224XRT3 A0A1I8QB48 A0A1J1HQF7 A0A1B0FAE1 A0A0A1WFX9 W5JDZ2 A0A034VKC8 A0A3R7PW09 A0A2M3Z542 A0A182FJ28 A0A0P4VWQ0 T1HZ76 A0A067RDZ0 A0A1B0CHN3 A0A0L0C7Z7 A0A2R7VWL4 A0A1L8DDU9 A0A0P6HIW7 A0A0P5HU50 D6X3C6 A0A2M4BME2 A0A2M4BMD4 A0A0A9XPU8 A0A2M4BN94 E2A8A7 A0A0L7QT22 A0A146KTF9 A0A0K8WAK3 A0A0P5NMU8
A0A087ZSM6 A0A2A3EEM6 A0A2P8Y3U1 E2BCI5 U4UQ50 N6UMK4 B0W9K6 A0A1Y1MH74 A0A0T6B3K2 B4L2V1 A0A1Q3F8C4 A0A1Q3F8A1 A0A1Q3F8G2 A0A195C287 A0A182JIP4 A0A182N1A8 A0A182JSU1 A0A3L8E179 B4JWR7 A0A182W4N4 U5ENR4 A0A182SMI5 A0A182YMY7 A0A084VX09 B4M379 A0A182RBI4 A0A151IR96 A0A1B6ISH0 A0A026WY37 A0A1B6KR95 A0A182XLT3 F4WUF7 A0A182IA91 A0A1S4G957 A0A158NN40 A0A1B6EKM7 B4ILC9 X2JLL7 Q9VR50 A0A182USS5 A0A151X0C3 A0A1A9X8T0 B3NYG8 B4N1K1 A0A1I8MKA0 Q7QFK8 A0A0M4EJT5 B4PZ64 Q29JF1 B4H301 A0A151HYP1 A0A182Q475 A0A182TUW8 A0A1A9Z2E3 A0A1A9VRL1 A0A023ER00 A0A1W4VWR4 B3MRT5 A0A3B0J6N3 A0A182HEA2 A0A1A9WCB4 A0A151JVG2 A0A182P7W1 A0A0K8VZ88 A0A224XRT3 A0A1I8QB48 A0A1J1HQF7 A0A1B0FAE1 A0A0A1WFX9 W5JDZ2 A0A034VKC8 A0A3R7PW09 A0A2M3Z542 A0A182FJ28 A0A0P4VWQ0 T1HZ76 A0A067RDZ0 A0A1B0CHN3 A0A0L0C7Z7 A0A2R7VWL4 A0A1L8DDU9 A0A0P6HIW7 A0A0P5HU50 D6X3C6 A0A2M4BME2 A0A2M4BMD4 A0A0A9XPU8 A0A2M4BN94 E2A8A7 A0A0L7QT22 A0A146KTF9 A0A0K8WAK3 A0A0P5NMU8
PDB
5Y79
E-value=0.000944305,
Score=99
Ontologies
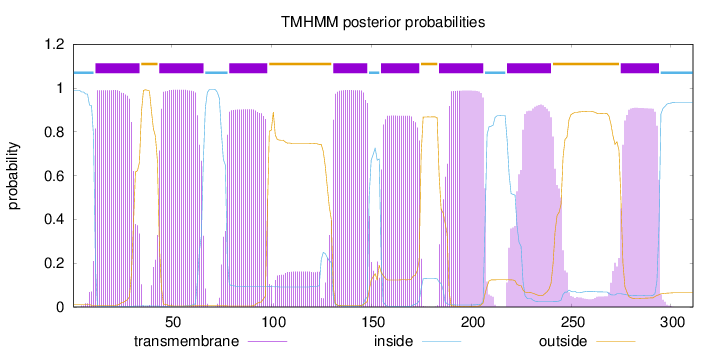
Topology
Subcellular location
Membrane
Length:
311
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
162.87889
Exp number, first 60 AAs:
37.88398
Total prob of N-in:
0.98916
POSSIBLE N-term signal
sequence
inside
1 - 11
TMhelix
12 - 34
outside
35 - 43
TMhelix
44 - 66
inside
67 - 78
TMhelix
79 - 98
outside
99 - 130
TMhelix
131 - 148
inside
149 - 154
TMhelix
155 - 174
outside
175 - 183
TMhelix
184 - 206
inside
207 - 217
TMhelix
218 - 240
outside
241 - 274
TMhelix
275 - 294
inside
295 - 311
Population Genetic Test Statistics
Pi
224.426852
Theta
180.430378
Tajima's D
0.92677
CLR
0
CSRT
0.642617869106545
Interpretation
Uncertain
