Gene
KWMTBOMO12836
Pre Gene Modal
BGIBMGA012748
Annotation
PREDICTED:_lachesin_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.865
Sequence
CDS
ATGGTCGAGCCTGATGCGAGCATGGCGTGGAGACGGGGGCCCTGCAAAAGCTTCGCCATCAGCGTCATACAGATCATCACTATTATATGTCAAGTGTTAACAGAGGAGCCTCGGTTTGCTGAGCCCATCCCTAACGTGACCGTGGCTCTGGGCCGAGATGCCAGCCTCCCGTGTGTCGTGGAACATCTCGGGACCTATAAGGTCGCCTGGATCCACATCGACCGCCAGATGATCCTGACCATCCATCGTCATGTCATCACTCGCCTGGCCCGGTTCAGCGTGTCGCATGACAACGCCATGACCTGGCTGCTCCACGTCAGCCAGGTCCAGCAAGAAGACAGAGGATATTACATGTGCCAAGTGAACACCAACCCTATGATCAGCCAAGTCGGATACCTTCAGGTTGTAGTTCCTCCAAATATCCTGGACGAAAAAAGTACGCAGTCAGCTGTTGCGGTGAGAGAAAACCAGAACATCAGCTTGGTGTGCAAGGCTGATGGATTTCCTACGCCCAAGATCATGTGGCGACGGGAGGACGGACAACCGATATCTGTGGACAGAAGGAAGAAAGTGACAGTGTACGAAGGTGAAATGCTGAATCTTCAACGCATAAGTCGCACGGAAATGGGCGCGTACCTCTGCATTGCTACCAACGCTGTGCCTCCGTCTGTCTCCAAGAGGATTATTGTGGACGTTGAGTTTTCACCCATGATCTGGGTACCGAATCAACTAGTGGGTGCTCCGGCCGGCACCGACGTGACGGTGGATTGCCACACGGAGGCCCACCCCCGCGCCATCTCCTACTGGGTCTATGATAACGTAATGGTACTTCCGACTAAGAAGTACGCGATCAATACTGAAGAAAATTCGTACAGGGCCCACATGAAGCTAACAGTGCGGAGTCTGCAGCTCGGCGACTTTGGCAACTACAGGTGCATATCGAAGAACTCATTAGGAGAGACTGAAGGTTCTATAAGGCTGTACGAAATTCCTATGCCATCGTCTTCACCAAAAGCCACAGAAATGAAGAGCAATGCCAACAAAGAAAGTCCGCGGCGTGTAAACGCGAGCCGGTCGGTCCCCCGCGCGGACTCGAACACGGAGCGGCCGAGCGTGGTCCGCGCGCAGCTCGACCGCTCGCCGGAGCGCGACGCCGCCCAGCACGTGTACCGCCCGCCGCACACGCACCACGTCTCAGGAACGAAAGCAGTGCATTGTTGGAGACCTTCAGTATTAGCTTTTTTTGTCTTATTTCAAATGGATTTTGTAGCCGATTTTTTTTGA
Protein
MVEPDASMAWRRGPCKSFAISVIQIITIICQVLTEEPRFAEPIPNVTVALGRDASLPCVVEHLGTYKVAWIHIDRQMILTIHRHVITRLARFSVSHDNAMTWLLHVSQVQQEDRGYYMCQVNTNPMISQVGYLQVVVPPNILDEKSTQSAVAVRENQNISLVCKADGFPTPKIMWRREDGQPISVDRRKKVTVYEGEMLNLQRISRTEMGAYLCIATNAVPPSVSKRIIVDVEFSPMIWVPNQLVGAPAGTDVTVDCHTEAHPRAISYWVYDNVMVLPTKKYAINTEENSYRAHMKLTVRSLQLGDFGNYRCISKNSLGETEGSIRLYEIPMPSSSPKATEMKSNANKESPRRVNASRSVPRADSNTERPSVVRAQLDRSPERDAAQHVYRPPHTHHVSGTKAVHCWRPSVLAFFVLFQMDFVADFF
Summary
Uniprot
A0A2W1BNC4
A0A2A4JWV6
A0A212F4T6
A0A0N0PCF9
A0A194Q814
D6W6G4
+ More
A0A0T6AXE6 A0A1B6KD26 A0A1J1HGP7 E0VR02 A0A182IJ74 A0A084WCV3 A0A182V3N6 A0A182HQH7 A0A182LDV7 Q7Q807 A0A182YKP7 A0A2M4DQC5 A0A182PWI3 W5J9W3 A0A182QHC1 A0A1I8Q0H9 W8BUY3 A0A1A9Y9P1 A0A1B0C2D3 A0A023F435 A0A1S4G204 A0A224XMS5 A0A182N218 Q16GM1 A0A182FEP8 Q17KA9 A0A0M4EYB4 A0A088ANS3 B4LBD0 B4KVC0 A0A0M4EJH0 A0A182H8J8 A0A0R3P4G9 B5DPC0 A0A3B0J9A5 A0A1W4VSI4 A0A0Q9WZ38 A0A0P9C470 A0A0Q5UIC3 A0A0J9RP80 A8JNC7 A0A0R1DZ80 A0A232FI53 A0A182WLY4 K7IX25 A0A0Q5UIF2 A0A0R1E0W0 A0A158P138 A0A1B0A2E6 A0A1B0CCX4 Q1RKT2 A0A2S2QN20 J9KD52 A0A2S2NXN2 A0A1A9X2X6 A0A0L0CBW8 A0A336MB36 A0A336LSY5 A0A1W4WQ57 X1WJ58 C4WUZ3 A0A2H8TNY1 A0A2S2Q8N6 A0A151XK58 A0A2S2NQW2 T1IJH4 T1IZV8 T1IWH8 A0A1Y1KSH7 A0A1B6JAN5 A0A1B6H1T5 A0A1B6MB79 E9FUV1 T1DFY0 A0A1B6ENA4 D6W8U0 A0A139WMQ5
A0A0T6AXE6 A0A1B6KD26 A0A1J1HGP7 E0VR02 A0A182IJ74 A0A084WCV3 A0A182V3N6 A0A182HQH7 A0A182LDV7 Q7Q807 A0A182YKP7 A0A2M4DQC5 A0A182PWI3 W5J9W3 A0A182QHC1 A0A1I8Q0H9 W8BUY3 A0A1A9Y9P1 A0A1B0C2D3 A0A023F435 A0A1S4G204 A0A224XMS5 A0A182N218 Q16GM1 A0A182FEP8 Q17KA9 A0A0M4EYB4 A0A088ANS3 B4LBD0 B4KVC0 A0A0M4EJH0 A0A182H8J8 A0A0R3P4G9 B5DPC0 A0A3B0J9A5 A0A1W4VSI4 A0A0Q9WZ38 A0A0P9C470 A0A0Q5UIC3 A0A0J9RP80 A8JNC7 A0A0R1DZ80 A0A232FI53 A0A182WLY4 K7IX25 A0A0Q5UIF2 A0A0R1E0W0 A0A158P138 A0A1B0A2E6 A0A1B0CCX4 Q1RKT2 A0A2S2QN20 J9KD52 A0A2S2NXN2 A0A1A9X2X6 A0A0L0CBW8 A0A336MB36 A0A336LSY5 A0A1W4WQ57 X1WJ58 C4WUZ3 A0A2H8TNY1 A0A2S2Q8N6 A0A151XK58 A0A2S2NQW2 T1IJH4 T1IZV8 T1IWH8 A0A1Y1KSH7 A0A1B6JAN5 A0A1B6H1T5 A0A1B6MB79 E9FUV1 T1DFY0 A0A1B6ENA4 D6W8U0 A0A139WMQ5
Pubmed
28756777
22118469
26354079
18362917
19820115
20566863
+ More
24438588 20966253 12364791 14747013 17210077 25244985 20920257 23761445 24495485 25474469 17510324 17994087 26483478 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 28648823 20075255 21347285 26108605 28004739 21292972
24438588 20966253 12364791 14747013 17210077 25244985 20920257 23761445 24495485 25474469 17510324 17994087 26483478 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 28648823 20075255 21347285 26108605 28004739 21292972
EMBL
KZ149953
PZC76562.1
NWSH01000472
PCG76188.1
AGBW02010320
OWR48733.1
+ More
KQ460557 KPJ13964.1 KQ459324 KPJ01663.1 KQ971307 EFA11064.2 LJIG01022641 KRT79486.1 GEBQ01030625 JAT09352.1 CVRI01000004 CRK87173.1 DS235442 EEB15808.1 ATLV01022886 ATLV01022887 KE525337 KFB48047.1 APCN01004610 AAAB01008948 EAA10490.4 GGFL01015575 MBW79753.1 ADMH02001733 ETN61252.1 AXCN02002351 GAMC01003568 JAC02988.1 JXJN01024493 JXJN01024494 GBBI01002491 JAC16221.1 GFTR01005298 JAW11128.1 CH478262 EAT33391.1 CH477227 EAT47076.1 CP012525 ALC43132.1 CH940647 EDW69718.2 CH933809 EDW18363.2 ALC43135.1 JXUM01029178 JXUM01029179 JXUM01029180 JXUM01029181 KQ560844 KXJ80649.1 CH379069 KRT08056.1 EDY73659.2 OUUW01000002 SPP76923.1 CH963847 KRF97676.1 CH902618 KPU78517.1 CH954178 KQS43638.1 CM002912 KMY97736.1 AE014296 ABW08467.2 AGB94136.1 CM000159 KRK01235.1 NNAY01000161 OXU30426.1 AAZX01006816 KQS43639.1 KRK01234.1 ADTU01005502 ADTU01005503 ADTU01005504 ADTU01005505 ADTU01005506 ADTU01005507 ADTU01005508 ADTU01005509 ADTU01005510 ADTU01005511 AJWK01007166 AJWK01007167 AJWK01007168 BT025128 ABE73299.1 GGMS01009870 MBY79073.1 ABLF02036987 ABLF02036989 ABLF02036990 ABLF02036993 ABLF02036998 ABLF02036999 ABLF02037001 ABLF02037004 ABLF02037006 GGMR01009203 MBY21822.1 JRES01000611 KNC29888.1 UFQT01000683 SSX26541.1 UFQS01000164 UFQT01000164 SSX00660.1 SSX21040.1 ABLF02030099 ABLF02040408 ABLF02040411 ABLF02040429 ABLF02040437 ABLF02040440 AK341286 BAH71713.1 GFXV01003826 MBW15631.1 GGMS01004788 MBY73991.1 KQ982036 KYQ60802.1 GGMR01006925 MBY19544.1 JH430290 JH431728 JH431617 GEZM01074983 JAV64359.1 GECU01011462 JAS96244.1 GECZ01001126 JAS68643.1 GEBQ01006795 JAT33182.1 GL732525 EFX88857.1 GAKT01000086 JAA92976.1 GECZ01030393 JAS39376.1 KQ971312 EEZ98398.2 KYB29127.1
KQ460557 KPJ13964.1 KQ459324 KPJ01663.1 KQ971307 EFA11064.2 LJIG01022641 KRT79486.1 GEBQ01030625 JAT09352.1 CVRI01000004 CRK87173.1 DS235442 EEB15808.1 ATLV01022886 ATLV01022887 KE525337 KFB48047.1 APCN01004610 AAAB01008948 EAA10490.4 GGFL01015575 MBW79753.1 ADMH02001733 ETN61252.1 AXCN02002351 GAMC01003568 JAC02988.1 JXJN01024493 JXJN01024494 GBBI01002491 JAC16221.1 GFTR01005298 JAW11128.1 CH478262 EAT33391.1 CH477227 EAT47076.1 CP012525 ALC43132.1 CH940647 EDW69718.2 CH933809 EDW18363.2 ALC43135.1 JXUM01029178 JXUM01029179 JXUM01029180 JXUM01029181 KQ560844 KXJ80649.1 CH379069 KRT08056.1 EDY73659.2 OUUW01000002 SPP76923.1 CH963847 KRF97676.1 CH902618 KPU78517.1 CH954178 KQS43638.1 CM002912 KMY97736.1 AE014296 ABW08467.2 AGB94136.1 CM000159 KRK01235.1 NNAY01000161 OXU30426.1 AAZX01006816 KQS43639.1 KRK01234.1 ADTU01005502 ADTU01005503 ADTU01005504 ADTU01005505 ADTU01005506 ADTU01005507 ADTU01005508 ADTU01005509 ADTU01005510 ADTU01005511 AJWK01007166 AJWK01007167 AJWK01007168 BT025128 ABE73299.1 GGMS01009870 MBY79073.1 ABLF02036987 ABLF02036989 ABLF02036990 ABLF02036993 ABLF02036998 ABLF02036999 ABLF02037001 ABLF02037004 ABLF02037006 GGMR01009203 MBY21822.1 JRES01000611 KNC29888.1 UFQT01000683 SSX26541.1 UFQS01000164 UFQT01000164 SSX00660.1 SSX21040.1 ABLF02030099 ABLF02040408 ABLF02040411 ABLF02040429 ABLF02040437 ABLF02040440 AK341286 BAH71713.1 GFXV01003826 MBW15631.1 GGMS01004788 MBY73991.1 KQ982036 KYQ60802.1 GGMR01006925 MBY19544.1 JH430290 JH431728 JH431617 GEZM01074983 JAV64359.1 GECU01011462 JAS96244.1 GECZ01001126 JAS68643.1 GEBQ01006795 JAT33182.1 GL732525 EFX88857.1 GAKT01000086 JAA92976.1 GECZ01030393 JAS39376.1 KQ971312 EEZ98398.2 KYB29127.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000053268
UP000007266
UP000183832
+ More
UP000009046 UP000075880 UP000030765 UP000075903 UP000075840 UP000075882 UP000007062 UP000076408 UP000075885 UP000000673 UP000075886 UP000095300 UP000092443 UP000092460 UP000075884 UP000008820 UP000069272 UP000092553 UP000005203 UP000008792 UP000009192 UP000069940 UP000249989 UP000001819 UP000268350 UP000192221 UP000007798 UP000007801 UP000008711 UP000000803 UP000002282 UP000215335 UP000075920 UP000002358 UP000005205 UP000092445 UP000092461 UP000007819 UP000091820 UP000037069 UP000192223 UP000075809 UP000000305
UP000009046 UP000075880 UP000030765 UP000075903 UP000075840 UP000075882 UP000007062 UP000076408 UP000075885 UP000000673 UP000075886 UP000095300 UP000092443 UP000092460 UP000075884 UP000008820 UP000069272 UP000092553 UP000005203 UP000008792 UP000009192 UP000069940 UP000249989 UP000001819 UP000268350 UP000192221 UP000007798 UP000007801 UP000008711 UP000000803 UP000002282 UP000215335 UP000075920 UP000002358 UP000005205 UP000092445 UP000092461 UP000007819 UP000091820 UP000037069 UP000192223 UP000075809 UP000000305
PRIDE
Interpro
SUPFAM
SSF48726
SSF48726
Gene 3D
ProteinModelPortal
A0A2W1BNC4
A0A2A4JWV6
A0A212F4T6
A0A0N0PCF9
A0A194Q814
D6W6G4
+ More
A0A0T6AXE6 A0A1B6KD26 A0A1J1HGP7 E0VR02 A0A182IJ74 A0A084WCV3 A0A182V3N6 A0A182HQH7 A0A182LDV7 Q7Q807 A0A182YKP7 A0A2M4DQC5 A0A182PWI3 W5J9W3 A0A182QHC1 A0A1I8Q0H9 W8BUY3 A0A1A9Y9P1 A0A1B0C2D3 A0A023F435 A0A1S4G204 A0A224XMS5 A0A182N218 Q16GM1 A0A182FEP8 Q17KA9 A0A0M4EYB4 A0A088ANS3 B4LBD0 B4KVC0 A0A0M4EJH0 A0A182H8J8 A0A0R3P4G9 B5DPC0 A0A3B0J9A5 A0A1W4VSI4 A0A0Q9WZ38 A0A0P9C470 A0A0Q5UIC3 A0A0J9RP80 A8JNC7 A0A0R1DZ80 A0A232FI53 A0A182WLY4 K7IX25 A0A0Q5UIF2 A0A0R1E0W0 A0A158P138 A0A1B0A2E6 A0A1B0CCX4 Q1RKT2 A0A2S2QN20 J9KD52 A0A2S2NXN2 A0A1A9X2X6 A0A0L0CBW8 A0A336MB36 A0A336LSY5 A0A1W4WQ57 X1WJ58 C4WUZ3 A0A2H8TNY1 A0A2S2Q8N6 A0A151XK58 A0A2S2NQW2 T1IJH4 T1IZV8 T1IWH8 A0A1Y1KSH7 A0A1B6JAN5 A0A1B6H1T5 A0A1B6MB79 E9FUV1 T1DFY0 A0A1B6ENA4 D6W8U0 A0A139WMQ5
A0A0T6AXE6 A0A1B6KD26 A0A1J1HGP7 E0VR02 A0A182IJ74 A0A084WCV3 A0A182V3N6 A0A182HQH7 A0A182LDV7 Q7Q807 A0A182YKP7 A0A2M4DQC5 A0A182PWI3 W5J9W3 A0A182QHC1 A0A1I8Q0H9 W8BUY3 A0A1A9Y9P1 A0A1B0C2D3 A0A023F435 A0A1S4G204 A0A224XMS5 A0A182N218 Q16GM1 A0A182FEP8 Q17KA9 A0A0M4EYB4 A0A088ANS3 B4LBD0 B4KVC0 A0A0M4EJH0 A0A182H8J8 A0A0R3P4G9 B5DPC0 A0A3B0J9A5 A0A1W4VSI4 A0A0Q9WZ38 A0A0P9C470 A0A0Q5UIC3 A0A0J9RP80 A8JNC7 A0A0R1DZ80 A0A232FI53 A0A182WLY4 K7IX25 A0A0Q5UIF2 A0A0R1E0W0 A0A158P138 A0A1B0A2E6 A0A1B0CCX4 Q1RKT2 A0A2S2QN20 J9KD52 A0A2S2NXN2 A0A1A9X2X6 A0A0L0CBW8 A0A336MB36 A0A336LSY5 A0A1W4WQ57 X1WJ58 C4WUZ3 A0A2H8TNY1 A0A2S2Q8N6 A0A151XK58 A0A2S2NQW2 T1IJH4 T1IZV8 T1IWH8 A0A1Y1KSH7 A0A1B6JAN5 A0A1B6H1T5 A0A1B6MB79 E9FUV1 T1DFY0 A0A1B6ENA4 D6W8U0 A0A139WMQ5
PDB
6EG1
E-value=7.575e-88,
Score=826
Ontologies
GO
Topology
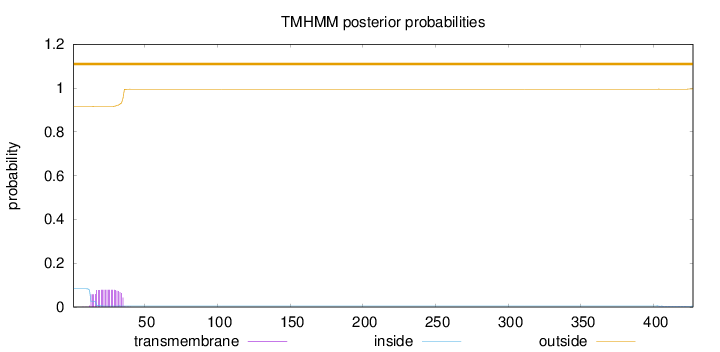
Length:
427
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.70558
Exp number, first 60 AAs:
1.66218
Total prob of N-in:
0.08342
outside
1 - 427
Population Genetic Test Statistics
Pi
228.186241
Theta
178.601063
Tajima's D
0.561017
CLR
0.345097
CSRT
0.536773161341933
Interpretation
Uncertain
