Gene
KWMTBOMO12831
Pre Gene Modal
BGIBMGA012730
Annotation
neuropeptide_receptor_A12_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.727
Sequence
CDS
ATGACAGTTCTTAGGATGGAGGCACAGCTCTTCAAGAAGGCTGGTTGTGTCACAGATATATTCCTGGTGAATCTGTCGGCGGCAGACCTACTCGTGACTGGCATTTGTATGCCGATCCAACTGAGCAAGGCCATTACCCTCGTTTGGTTCTACGGAGAGACTGTTTGCAAGATTGTTAATTACATTCAAGGCGTGGCTGTGGCGGCTAGCGTGTTCACGATCTCTGCGATGTCAGTGGACCGTTGGTTGTCCATTACTCCAGAACCTCGTCTTCGGCCACCAGGCAGGAAGCAGGCGACGCTGCTCCTCATGCTACTTTGGATCGCTGCACTGCTTATCTTCATTCCAACCTCTCTAGTAGCAGGAGTGCGGAAGGAAACCATTCCTATAATATCCAAAGGAGATAAGAACATTTCAATAGAAACCAGAGATATCCATTTCTGCATTGAGGAATGGCCTTCACCGGAGACCAGGAAGCAGTATGGAATGTTCAGTTTCACTCTAGTCTACGCAATACCAGGTCAGAATTAA
Protein
MTVLRMEAQLFKKAGCVTDIFLVNLSAADLLVTGICMPIQLSKAITLVWFYGETVCKIVNYIQGVAVAASVFTISAMSVDRWLSITPEPRLRPPGRKQATLLLMLLWIAALLIFIPTSLVAGVRKETIPIISKGDKNISIETRDIHFCIEEWPSPETRKQYGMFSFTLVYAIPGQN
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
B3XXM5
A0A2H1VED4
A0A0S1YDB4
A0A2A4J221
A0A194RBD9
A0A212EH34
+ More
A0A2W1BZI0 A0A194Q809 A0A212FL87 A0A194QDW6 A0A437B4D2 B3XXP8 A0A2W1BT88 A0A0S1YD95 A0A1B6CDE5 A0A2H8TQ41 J9LMX5 A0A2S2NN60 A0A2S2QWP3 A0A087ZYM9 A0A2A3EGD0 A0A226EKJ1 V4ALY3 V4ASX2 A0A0M8ZPP4 A0A1W4XAT5 A0A1W4WZW7 A0A154PQW2 A0A1W4WZV7 A0A1Y1NH52 A0A3L8D745 A0A232FB03 A0A0N8BH82 A0A0K0PUJ7 A0A0K0PUI1 V4ALZ8 A0A210QBI3 E2BMY8 A0A452THC1 A0A3Q7XS02 A0A0P6DPN9 A0A0N8DJF0 X1Z504 A0A3Q7QAK7 A0A2U3YAH7 A0A2Y9I1X3 A0A310SGA3 D2I5D3 A0A2Y9JN69 A0A2K6LIW2 A0A2K6P4H4 A0A384C9N2 A0A452Q8V7 V4BH55 A0A2U3ZLS7 A0A2T7NRV4 R7V1S8 A0A3P4PZP4 I3MS29 A0A2K6LIW1 A0A2K6P4H8 L5LHS2 G1PAR9 F7EXM8 A0A2K6DQR3 A0A2K6DQS1 A0A2K5W108 F7A1W4 A0A2K5MJY8 M3YD64 A0A2K5MJZ4 A0A0D9QXR6 G3WBN9 A0A2K5I6Y5 A0A096N0F7 A0A2I3LG05 S7Q1T7 A0A2K5YFV8 A0A2K5YFU6 M3WWS8 A0A1L8HVP2 A0A1B8Y8P3 A0A091IRM8 F6ZDD4 D6WTB3 A0A2R9AVQ6 A0A2I3TM59 A0A2K6DQQ6 A0A2K5W196 A0A2J8PFD3 A0A2J8PFC6 A0A139WFF0
A0A2W1BZI0 A0A194Q809 A0A212FL87 A0A194QDW6 A0A437B4D2 B3XXP8 A0A2W1BT88 A0A0S1YD95 A0A1B6CDE5 A0A2H8TQ41 J9LMX5 A0A2S2NN60 A0A2S2QWP3 A0A087ZYM9 A0A2A3EGD0 A0A226EKJ1 V4ALY3 V4ASX2 A0A0M8ZPP4 A0A1W4XAT5 A0A1W4WZW7 A0A154PQW2 A0A1W4WZV7 A0A1Y1NH52 A0A3L8D745 A0A232FB03 A0A0N8BH82 A0A0K0PUJ7 A0A0K0PUI1 V4ALZ8 A0A210QBI3 E2BMY8 A0A452THC1 A0A3Q7XS02 A0A0P6DPN9 A0A0N8DJF0 X1Z504 A0A3Q7QAK7 A0A2U3YAH7 A0A2Y9I1X3 A0A310SGA3 D2I5D3 A0A2Y9JN69 A0A2K6LIW2 A0A2K6P4H4 A0A384C9N2 A0A452Q8V7 V4BH55 A0A2U3ZLS7 A0A2T7NRV4 R7V1S8 A0A3P4PZP4 I3MS29 A0A2K6LIW1 A0A2K6P4H8 L5LHS2 G1PAR9 F7EXM8 A0A2K6DQR3 A0A2K6DQS1 A0A2K5W108 F7A1W4 A0A2K5MJY8 M3YD64 A0A2K5MJZ4 A0A0D9QXR6 G3WBN9 A0A2K5I6Y5 A0A096N0F7 A0A2I3LG05 S7Q1T7 A0A2K5YFV8 A0A2K5YFU6 M3WWS8 A0A1L8HVP2 A0A1B8Y8P3 A0A091IRM8 F6ZDD4 D6WTB3 A0A2R9AVQ6 A0A2I3TM59 A0A2K6DQQ6 A0A2K5W196 A0A2J8PFD3 A0A2J8PFC6 A0A139WFF0
Pubmed
EMBL
AB330433
BAG68411.1
ODYU01001854
SOQ38634.1
KT031010
ALM88308.1
+ More
NWSH01003879 PCG65728.1 KQ460416 KPJ14932.1 AGBW02014980 OWR40781.1 KZ149907 PZC78270.1 KQ459324 KPJ01658.1 AGBW02007828 OWR54496.1 KPJ01656.1 RSAL01000176 RVE45033.1 AB330456 BAG68434.1 PZC78269.1 KT031033 ALM88331.1 GEDC01025821 JAS11477.1 GFXV01004314 MBW16119.1 ABLF02017913 ABLF02017914 ABLF02017915 GGMR01006004 MBY18623.1 GGMS01012349 MBY81552.1 KZ288262 PBC30352.1 LNIX01000003 OXA57810.1 KB201802 ESO94606.1 KB200750 ESP00348.1 KQ435925 KOX68387.1 KQ435037 KZC14127.1 GEZM01002875 JAV97099.1 QOIP01000012 RLU16307.1 NNAY01000533 OXU27835.1 GDIQ01178003 JAK73722.1 KP294010 AKQ63064.1 KP293979 AKQ63033.1 KB199650 ESP05214.1 NEDP02004282 OWF46089.1 GL449382 EFN82892.1 GDIQ01074229 JAN20508.1 GDIP01027847 LRGB01000868 JAM75868.1 KZS15562.1 AMQN01000412 KQ760243 OAD61277.1 ACTA01068273 GL194651 EFB21559.1 ESP05282.1 PZQS01000010 PVD23914.1 AMQN01006107 KB297637 ELU10276.1 CYRY02035320 VCX15563.1 AGTP01072032 KB111812 ELK25535.1 AAPE02028687 JSUE03033561 JSUE03033562 JSUE03033563 AQIA01054851 AQIA01054852 AQIA01054853 AEYP01065790 AEYP01065791 AEYP01065792 AEYP01065793 AEYP01065794 AQIB01016335 AEFK01211418 AEFK01211419 AEFK01211420 AHZZ02026895 AHZZ02026896 AHZZ02026897 AHZZ02026898 AHZZ02026899 KE164343 EPQ17278.1 AANG04001605 CM004466 OCU00058.1 KV460381 OCA19342.1 KK500905 KFP11354.1 AAMC01103204 AAMC01103205 KQ971352 EFA07466.2 AJFE02062850 AJFE02062851 AJFE02062852 AJFE02062853 AACZ04033881 NBAG03000215 PNI82722.1 PNI82721.1 KYB26646.1
NWSH01003879 PCG65728.1 KQ460416 KPJ14932.1 AGBW02014980 OWR40781.1 KZ149907 PZC78270.1 KQ459324 KPJ01658.1 AGBW02007828 OWR54496.1 KPJ01656.1 RSAL01000176 RVE45033.1 AB330456 BAG68434.1 PZC78269.1 KT031033 ALM88331.1 GEDC01025821 JAS11477.1 GFXV01004314 MBW16119.1 ABLF02017913 ABLF02017914 ABLF02017915 GGMR01006004 MBY18623.1 GGMS01012349 MBY81552.1 KZ288262 PBC30352.1 LNIX01000003 OXA57810.1 KB201802 ESO94606.1 KB200750 ESP00348.1 KQ435925 KOX68387.1 KQ435037 KZC14127.1 GEZM01002875 JAV97099.1 QOIP01000012 RLU16307.1 NNAY01000533 OXU27835.1 GDIQ01178003 JAK73722.1 KP294010 AKQ63064.1 KP293979 AKQ63033.1 KB199650 ESP05214.1 NEDP02004282 OWF46089.1 GL449382 EFN82892.1 GDIQ01074229 JAN20508.1 GDIP01027847 LRGB01000868 JAM75868.1 KZS15562.1 AMQN01000412 KQ760243 OAD61277.1 ACTA01068273 GL194651 EFB21559.1 ESP05282.1 PZQS01000010 PVD23914.1 AMQN01006107 KB297637 ELU10276.1 CYRY02035320 VCX15563.1 AGTP01072032 KB111812 ELK25535.1 AAPE02028687 JSUE03033561 JSUE03033562 JSUE03033563 AQIA01054851 AQIA01054852 AQIA01054853 AEYP01065790 AEYP01065791 AEYP01065792 AEYP01065793 AEYP01065794 AQIB01016335 AEFK01211418 AEFK01211419 AEFK01211420 AHZZ02026895 AHZZ02026896 AHZZ02026897 AHZZ02026898 AHZZ02026899 KE164343 EPQ17278.1 AANG04001605 CM004466 OCU00058.1 KV460381 OCA19342.1 KK500905 KFP11354.1 AAMC01103204 AAMC01103205 KQ971352 EFA07466.2 AJFE02062850 AJFE02062851 AJFE02062852 AJFE02062853 AACZ04033881 NBAG03000215 PNI82722.1 PNI82721.1 KYB26646.1
Proteomes
UP000218220
UP000053240
UP000007151
UP000053268
UP000283053
UP000007819
+ More
UP000005203 UP000242457 UP000198287 UP000030746 UP000053105 UP000192223 UP000076502 UP000279307 UP000215335 UP000242188 UP000008237 UP000291021 UP000286642 UP000076858 UP000286641 UP000245341 UP000248481 UP000008912 UP000248482 UP000233180 UP000233200 UP000261680 UP000291022 UP000245340 UP000245119 UP000014760 UP000005215 UP000001074 UP000006718 UP000233120 UP000233100 UP000233060 UP000000715 UP000029965 UP000007648 UP000233080 UP000028761 UP000233140 UP000011712 UP000186698 UP000008143 UP000053119 UP000007266 UP000240080 UP000002277
UP000005203 UP000242457 UP000198287 UP000030746 UP000053105 UP000192223 UP000076502 UP000279307 UP000215335 UP000242188 UP000008237 UP000291021 UP000286642 UP000076858 UP000286641 UP000245341 UP000248481 UP000008912 UP000248482 UP000233180 UP000233200 UP000261680 UP000291022 UP000245340 UP000245119 UP000014760 UP000005215 UP000001074 UP000006718 UP000233120 UP000233100 UP000233060 UP000000715 UP000029965 UP000007648 UP000233080 UP000028761 UP000233140 UP000011712 UP000186698 UP000008143 UP000053119 UP000007266 UP000240080 UP000002277
Pfam
PF00001 7tm_1
Interpro
ProteinModelPortal
B3XXM5
A0A2H1VED4
A0A0S1YDB4
A0A2A4J221
A0A194RBD9
A0A212EH34
+ More
A0A2W1BZI0 A0A194Q809 A0A212FL87 A0A194QDW6 A0A437B4D2 B3XXP8 A0A2W1BT88 A0A0S1YD95 A0A1B6CDE5 A0A2H8TQ41 J9LMX5 A0A2S2NN60 A0A2S2QWP3 A0A087ZYM9 A0A2A3EGD0 A0A226EKJ1 V4ALY3 V4ASX2 A0A0M8ZPP4 A0A1W4XAT5 A0A1W4WZW7 A0A154PQW2 A0A1W4WZV7 A0A1Y1NH52 A0A3L8D745 A0A232FB03 A0A0N8BH82 A0A0K0PUJ7 A0A0K0PUI1 V4ALZ8 A0A210QBI3 E2BMY8 A0A452THC1 A0A3Q7XS02 A0A0P6DPN9 A0A0N8DJF0 X1Z504 A0A3Q7QAK7 A0A2U3YAH7 A0A2Y9I1X3 A0A310SGA3 D2I5D3 A0A2Y9JN69 A0A2K6LIW2 A0A2K6P4H4 A0A384C9N2 A0A452Q8V7 V4BH55 A0A2U3ZLS7 A0A2T7NRV4 R7V1S8 A0A3P4PZP4 I3MS29 A0A2K6LIW1 A0A2K6P4H8 L5LHS2 G1PAR9 F7EXM8 A0A2K6DQR3 A0A2K6DQS1 A0A2K5W108 F7A1W4 A0A2K5MJY8 M3YD64 A0A2K5MJZ4 A0A0D9QXR6 G3WBN9 A0A2K5I6Y5 A0A096N0F7 A0A2I3LG05 S7Q1T7 A0A2K5YFV8 A0A2K5YFU6 M3WWS8 A0A1L8HVP2 A0A1B8Y8P3 A0A091IRM8 F6ZDD4 D6WTB3 A0A2R9AVQ6 A0A2I3TM59 A0A2K6DQQ6 A0A2K5W196 A0A2J8PFD3 A0A2J8PFC6 A0A139WFF0
A0A2W1BZI0 A0A194Q809 A0A212FL87 A0A194QDW6 A0A437B4D2 B3XXP8 A0A2W1BT88 A0A0S1YD95 A0A1B6CDE5 A0A2H8TQ41 J9LMX5 A0A2S2NN60 A0A2S2QWP3 A0A087ZYM9 A0A2A3EGD0 A0A226EKJ1 V4ALY3 V4ASX2 A0A0M8ZPP4 A0A1W4XAT5 A0A1W4WZW7 A0A154PQW2 A0A1W4WZV7 A0A1Y1NH52 A0A3L8D745 A0A232FB03 A0A0N8BH82 A0A0K0PUJ7 A0A0K0PUI1 V4ALZ8 A0A210QBI3 E2BMY8 A0A452THC1 A0A3Q7XS02 A0A0P6DPN9 A0A0N8DJF0 X1Z504 A0A3Q7QAK7 A0A2U3YAH7 A0A2Y9I1X3 A0A310SGA3 D2I5D3 A0A2Y9JN69 A0A2K6LIW2 A0A2K6P4H4 A0A384C9N2 A0A452Q8V7 V4BH55 A0A2U3ZLS7 A0A2T7NRV4 R7V1S8 A0A3P4PZP4 I3MS29 A0A2K6LIW1 A0A2K6P4H8 L5LHS2 G1PAR9 F7EXM8 A0A2K6DQR3 A0A2K6DQS1 A0A2K5W108 F7A1W4 A0A2K5MJY8 M3YD64 A0A2K5MJZ4 A0A0D9QXR6 G3WBN9 A0A2K5I6Y5 A0A096N0F7 A0A2I3LG05 S7Q1T7 A0A2K5YFV8 A0A2K5YFU6 M3WWS8 A0A1L8HVP2 A0A1B8Y8P3 A0A091IRM8 F6ZDD4 D6WTB3 A0A2R9AVQ6 A0A2I3TM59 A0A2K6DQQ6 A0A2K5W196 A0A2J8PFD3 A0A2J8PFC6 A0A139WFF0
PDB
5WS3
E-value=3.44075e-15,
Score=194
Ontologies
KEGG
GO
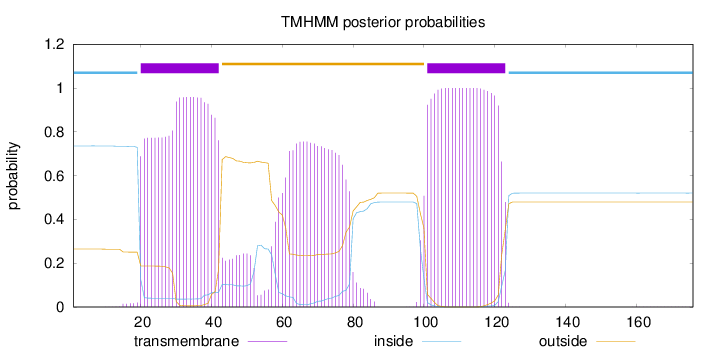
Topology
Length:
176
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
60.53373
Exp number, first 60 AAs:
23.97423
Total prob of N-in:
0.73566
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 42
outside
43 - 100
TMhelix
101 - 123
inside
124 - 176
Population Genetic Test Statistics
Pi
265.167388
Theta
172.516419
Tajima's D
1.464018
CLR
0.639921
CSRT
0.779061046947653
Interpretation
Uncertain
