Gene
KWMTBOMO12827
Pre Gene Modal
BGIBMGA012747
Annotation
hypothetical_protein_KGM_14827_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.442 Extracellular Reliability : 1.54
Sequence
CDS
ATGTCCGGTCTAAGTGTCGTCTTCAATGTGGAGCCCGGAGACTATCCGAGCTGGTCACCTATTCCTTATTACGGAGTCAAGGTCCTCATCAGTGACCCCAACGATTATCCGGAAACAACAGTGCTCTACCGTTTCATAACCTTAGGGGAATCAGTGGCCATCAAAGTTGACCCGATGGTTTTCCAGAGTGAGTCTAACATTCGCAGAGTGGTCCCCGACAAAAGGGCTTGCTGGTTTCACGATGAAGTGATCTTAGGGCACACTGACAGATATAGTTTCGAAACTTGTCACACCGAATGCAAGATGAAGACTTACTTGGATACTTGTGGATGTATCCCCTTTAAGTACCCAAGAGGTAAATACTTTTTTTAA
Protein
MSGLSVVFNVEPGDYPSWSPIPYYGVKVLISDPNDYPETTVLYRFITLGESVAIKVDPMVFQSESNIRRVVPDKRACWFHDEVILGHTDRYSFETCHTECKMKTYLDTCGCIPFKYPRGKYFF
Summary
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
H9JT85
A0A2W1BTN9
A0A2H1W7I2
A0A2A4J094
A0A212F4Q4
A0A0N0PAI0
+ More
A0A194Q8D8 A0A3S2P9C1 A0A2J7REJ0 A0A067RCE3 A0A1B0D5E9 A0A232F9M1 K7INU5 A0A336MTW7 A0A3Q0JDI0 A0A336M002 A0A1B0CK88 A0A195DYT3 A0A2P8YAC6 A0A0J7KTG4 A0A182S9T8 Q7PYW4 A0A182PN37 A0A182UX06 A0A1S4H745 A0A067RHK4 A0A151IQC2 A0A182LNY6 A0A2P8YAC3 A0A182WX49 A0A182HN58 A0A182UD57 A0A151WZQ4 A0A182Q221 A0A182YLS3 A0A139WIQ3 F4WCX3 A0A2J7QTA7 A0A151I1R2 A0A151IQ85 A0A195EWN8 A0A182IW77 E2A4M1 Q17NW3 A0A2J7QAD0 B0WDC4 A0A1B6HF37 A0A1S4EW59 A0A336KF41 A0A158NBB5 A0A182WIC2 A0A182KHA8 A0A182YNJ3 A0A182J6D0 A0A182R2G6 A0A182WEP1 A0A182X1C4 A0A182G9W4 A0A182SJN9 A0A158NWS2 E2BM03 A0A182W972 A0A195EVN0 A0A195DZ79 A0A182MLZ3 U4UHT6 A0A084WHX7 A0A0L7RDT9 N6UFK8 A0A1I8Q7F4 A0A182IDW7 A0A182QS19 A0A182MXN7 Q7Q066 A0A182UZ03 A0A182XCI7 A0A182THH2 A0A182MEP0 A0A182MVQ4 A0A182XD77 A0A182UXK6 A0A182JHD4 A0A182HQ19 A0A182MYI9 A0A182Y1U0 A0A182K171 A0A182U0T8 A0A182KSP4 A0A182N7Q2 A0A182GXU5 A0A0L0CPJ6 A0A154NXS5 Q7PF15 B0W821 A0A182GCI9 A0A182L5Y7 A0A1J1J775 Q16GX7 A0A1S4H2E9 Q7Q1X3
A0A194Q8D8 A0A3S2P9C1 A0A2J7REJ0 A0A067RCE3 A0A1B0D5E9 A0A232F9M1 K7INU5 A0A336MTW7 A0A3Q0JDI0 A0A336M002 A0A1B0CK88 A0A195DYT3 A0A2P8YAC6 A0A0J7KTG4 A0A182S9T8 Q7PYW4 A0A182PN37 A0A182UX06 A0A1S4H745 A0A067RHK4 A0A151IQC2 A0A182LNY6 A0A2P8YAC3 A0A182WX49 A0A182HN58 A0A182UD57 A0A151WZQ4 A0A182Q221 A0A182YLS3 A0A139WIQ3 F4WCX3 A0A2J7QTA7 A0A151I1R2 A0A151IQ85 A0A195EWN8 A0A182IW77 E2A4M1 Q17NW3 A0A2J7QAD0 B0WDC4 A0A1B6HF37 A0A1S4EW59 A0A336KF41 A0A158NBB5 A0A182WIC2 A0A182KHA8 A0A182YNJ3 A0A182J6D0 A0A182R2G6 A0A182WEP1 A0A182X1C4 A0A182G9W4 A0A182SJN9 A0A158NWS2 E2BM03 A0A182W972 A0A195EVN0 A0A195DZ79 A0A182MLZ3 U4UHT6 A0A084WHX7 A0A0L7RDT9 N6UFK8 A0A1I8Q7F4 A0A182IDW7 A0A182QS19 A0A182MXN7 Q7Q066 A0A182UZ03 A0A182XCI7 A0A182THH2 A0A182MEP0 A0A182MVQ4 A0A182XD77 A0A182UXK6 A0A182JHD4 A0A182HQ19 A0A182MYI9 A0A182Y1U0 A0A182K171 A0A182U0T8 A0A182KSP4 A0A182N7Q2 A0A182GXU5 A0A0L0CPJ6 A0A154NXS5 Q7PF15 B0W821 A0A182GCI9 A0A182L5Y7 A0A1J1J775 Q16GX7 A0A1S4H2E9 Q7Q1X3
Pubmed
EMBL
BABH01034832
KZ149953
PZC76567.1
ODYU01006832
SOQ49041.1
NWSH01004664
+ More
PCG64793.1 AGBW02010320 OWR48728.1 KQ458833 KPJ04917.1 KQ459324 KPJ01659.1 RSAL01000176 RVE45031.1 NEVH01004961 PNF39252.1 KK852549 KDR21541.1 AJVK01011749 NNAY01000660 OXU27138.1 UFQT01001968 SSX32233.1 UFQT01000343 SSX23390.1 AJWK01015896 AJWK01015897 KQ980050 KYN18028.1 PYGN01000758 PSN41205.1 LBMM01003440 KMQ93534.1 AAAB01008986 EAA00269.4 KK852504 KDR22518.1 KQ976782 KYN08320.1 PSN41206.1 APCN01001852 KQ982630 KYQ53387.1 AXCN02001053 KQ971338 KYB27860.1 GL888074 EGI67954.1 NEVH01011198 PNF31805.1 KQ976572 KYM80398.1 KYN08319.1 KQ981953 KYN32299.1 GL436710 EFN71623.1 CH477195 EAT48435.1 NEVH01016331 PNF25545.1 DS231896 EDS44437.1 GECU01034485 JAS73221.1 UFQS01000390 UFQT01000390 SSX03496.1 SSX23861.1 ADTU01010761 ADTU01010762 AXCP01006762 JXUM01050027 JXUM01050028 JXUM01050029 KQ561631 KXJ77947.1 ADTU01028435 GL449100 EFN83252.1 KYN32298.1 KYN18027.1 AXCM01007075 KB632364 ERL93559.1 ATLV01023896 KE525347 KFB49821.1 KQ414612 KOC69152.1 APGK01037684 KB740948 ENN77437.1 APCN01004535 AXCN02001041 EAA00234.4 AXCM01003932 AXCM01007400 APCN01003288 JXUM01096276 KQ564253 KXJ72524.1 JRES01000080 KNC34298.1 KQ434772 KZC03894.1 AAAB01008807 EAA45499.4 DS231856 EDS38483.1 JXUM01054263 KQ561813 KXJ77445.1 CVRI01000074 CRL08251.1 CH478228 EAT33496.1 AAAB01008980 EAA14549.4
PCG64793.1 AGBW02010320 OWR48728.1 KQ458833 KPJ04917.1 KQ459324 KPJ01659.1 RSAL01000176 RVE45031.1 NEVH01004961 PNF39252.1 KK852549 KDR21541.1 AJVK01011749 NNAY01000660 OXU27138.1 UFQT01001968 SSX32233.1 UFQT01000343 SSX23390.1 AJWK01015896 AJWK01015897 KQ980050 KYN18028.1 PYGN01000758 PSN41205.1 LBMM01003440 KMQ93534.1 AAAB01008986 EAA00269.4 KK852504 KDR22518.1 KQ976782 KYN08320.1 PSN41206.1 APCN01001852 KQ982630 KYQ53387.1 AXCN02001053 KQ971338 KYB27860.1 GL888074 EGI67954.1 NEVH01011198 PNF31805.1 KQ976572 KYM80398.1 KYN08319.1 KQ981953 KYN32299.1 GL436710 EFN71623.1 CH477195 EAT48435.1 NEVH01016331 PNF25545.1 DS231896 EDS44437.1 GECU01034485 JAS73221.1 UFQS01000390 UFQT01000390 SSX03496.1 SSX23861.1 ADTU01010761 ADTU01010762 AXCP01006762 JXUM01050027 JXUM01050028 JXUM01050029 KQ561631 KXJ77947.1 ADTU01028435 GL449100 EFN83252.1 KYN32298.1 KYN18027.1 AXCM01007075 KB632364 ERL93559.1 ATLV01023896 KE525347 KFB49821.1 KQ414612 KOC69152.1 APGK01037684 KB740948 ENN77437.1 APCN01004535 AXCN02001041 EAA00234.4 AXCM01003932 AXCM01007400 APCN01003288 JXUM01096276 KQ564253 KXJ72524.1 JRES01000080 KNC34298.1 KQ434772 KZC03894.1 AAAB01008807 EAA45499.4 DS231856 EDS38483.1 JXUM01054263 KQ561813 KXJ77445.1 CVRI01000074 CRL08251.1 CH478228 EAT33496.1 AAAB01008980 EAA14549.4
Proteomes
UP000005204
UP000218220
UP000007151
UP000053268
UP000283053
UP000235965
+ More
UP000027135 UP000092462 UP000215335 UP000002358 UP000079169 UP000092461 UP000078492 UP000245037 UP000036403 UP000075901 UP000007062 UP000075885 UP000075903 UP000078542 UP000075882 UP000076407 UP000075840 UP000075902 UP000075809 UP000075886 UP000076408 UP000007266 UP000007755 UP000078540 UP000078541 UP000075880 UP000000311 UP000008820 UP000002320 UP000005205 UP000075920 UP000075881 UP000075900 UP000069940 UP000249989 UP000008237 UP000075883 UP000030742 UP000030765 UP000053825 UP000019118 UP000095300 UP000075884 UP000037069 UP000076502 UP000183832
UP000027135 UP000092462 UP000215335 UP000002358 UP000079169 UP000092461 UP000078492 UP000245037 UP000036403 UP000075901 UP000007062 UP000075885 UP000075903 UP000078542 UP000075882 UP000076407 UP000075840 UP000075902 UP000075809 UP000075886 UP000076408 UP000007266 UP000007755 UP000078540 UP000078541 UP000075880 UP000000311 UP000008820 UP000002320 UP000005205 UP000075920 UP000075881 UP000075900 UP000069940 UP000249989 UP000008237 UP000075883 UP000030742 UP000030765 UP000053825 UP000019118 UP000095300 UP000075884 UP000037069 UP000076502 UP000183832
PRIDE
Interpro
SUPFAM
SSF54791
SSF54791
Gene 3D
ProteinModelPortal
H9JT85
A0A2W1BTN9
A0A2H1W7I2
A0A2A4J094
A0A212F4Q4
A0A0N0PAI0
+ More
A0A194Q8D8 A0A3S2P9C1 A0A2J7REJ0 A0A067RCE3 A0A1B0D5E9 A0A232F9M1 K7INU5 A0A336MTW7 A0A3Q0JDI0 A0A336M002 A0A1B0CK88 A0A195DYT3 A0A2P8YAC6 A0A0J7KTG4 A0A182S9T8 Q7PYW4 A0A182PN37 A0A182UX06 A0A1S4H745 A0A067RHK4 A0A151IQC2 A0A182LNY6 A0A2P8YAC3 A0A182WX49 A0A182HN58 A0A182UD57 A0A151WZQ4 A0A182Q221 A0A182YLS3 A0A139WIQ3 F4WCX3 A0A2J7QTA7 A0A151I1R2 A0A151IQ85 A0A195EWN8 A0A182IW77 E2A4M1 Q17NW3 A0A2J7QAD0 B0WDC4 A0A1B6HF37 A0A1S4EW59 A0A336KF41 A0A158NBB5 A0A182WIC2 A0A182KHA8 A0A182YNJ3 A0A182J6D0 A0A182R2G6 A0A182WEP1 A0A182X1C4 A0A182G9W4 A0A182SJN9 A0A158NWS2 E2BM03 A0A182W972 A0A195EVN0 A0A195DZ79 A0A182MLZ3 U4UHT6 A0A084WHX7 A0A0L7RDT9 N6UFK8 A0A1I8Q7F4 A0A182IDW7 A0A182QS19 A0A182MXN7 Q7Q066 A0A182UZ03 A0A182XCI7 A0A182THH2 A0A182MEP0 A0A182MVQ4 A0A182XD77 A0A182UXK6 A0A182JHD4 A0A182HQ19 A0A182MYI9 A0A182Y1U0 A0A182K171 A0A182U0T8 A0A182KSP4 A0A182N7Q2 A0A182GXU5 A0A0L0CPJ6 A0A154NXS5 Q7PF15 B0W821 A0A182GCI9 A0A182L5Y7 A0A1J1J775 Q16GX7 A0A1S4H2E9 Q7Q1X3
A0A194Q8D8 A0A3S2P9C1 A0A2J7REJ0 A0A067RCE3 A0A1B0D5E9 A0A232F9M1 K7INU5 A0A336MTW7 A0A3Q0JDI0 A0A336M002 A0A1B0CK88 A0A195DYT3 A0A2P8YAC6 A0A0J7KTG4 A0A182S9T8 Q7PYW4 A0A182PN37 A0A182UX06 A0A1S4H745 A0A067RHK4 A0A151IQC2 A0A182LNY6 A0A2P8YAC3 A0A182WX49 A0A182HN58 A0A182UD57 A0A151WZQ4 A0A182Q221 A0A182YLS3 A0A139WIQ3 F4WCX3 A0A2J7QTA7 A0A151I1R2 A0A151IQ85 A0A195EWN8 A0A182IW77 E2A4M1 Q17NW3 A0A2J7QAD0 B0WDC4 A0A1B6HF37 A0A1S4EW59 A0A336KF41 A0A158NBB5 A0A182WIC2 A0A182KHA8 A0A182YNJ3 A0A182J6D0 A0A182R2G6 A0A182WEP1 A0A182X1C4 A0A182G9W4 A0A182SJN9 A0A158NWS2 E2BM03 A0A182W972 A0A195EVN0 A0A195DZ79 A0A182MLZ3 U4UHT6 A0A084WHX7 A0A0L7RDT9 N6UFK8 A0A1I8Q7F4 A0A182IDW7 A0A182QS19 A0A182MXN7 Q7Q066 A0A182UZ03 A0A182XCI7 A0A182THH2 A0A182MEP0 A0A182MVQ4 A0A182XD77 A0A182UXK6 A0A182JHD4 A0A182HQ19 A0A182MYI9 A0A182Y1U0 A0A182K171 A0A182U0T8 A0A182KSP4 A0A182N7Q2 A0A182GXU5 A0A0L0CPJ6 A0A154NXS5 Q7PF15 B0W821 A0A182GCI9 A0A182L5Y7 A0A1J1J775 Q16GX7 A0A1S4H2E9 Q7Q1X3
Ontologies
KEGG
GO
PANTHER
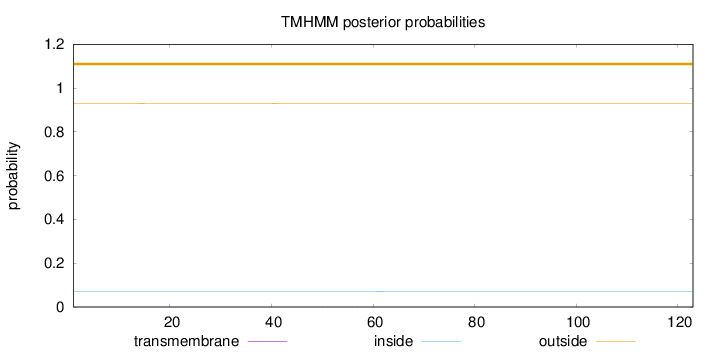
Topology
Length:
123
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00767
Exp number, first 60 AAs:
0.00734
Total prob of N-in:
0.07001
outside
1 - 123
Population Genetic Test Statistics
Pi
143.941135
Theta
175.038819
Tajima's D
0.704619
CLR
1.529754
CSRT
0.571021448927554
Interpretation
Uncertain
