Gene
KWMTBOMO12823
Annotation
PREDICTED:_uncharacterized_protein_LOC106708887_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 1.617 PlasmaMembrane Reliability : 1.581
Sequence
CDS
ATGTCGGTGTCCTACATTCGCGGGAAGGTGACTAGTGAGCTCCGATGGTTCGCATCAGCTCATGCTTTGGTTGACAGAACAGATAGGGATGCGGCATTGCTCGATAGGGCGTCTCAGGTGCACGGACGGTTGGAGGTAGTGTACGATCGGTTGTCTGACCTCGTTGCTTTGGGGTCCGAAGTGGAGAAGGAGGTGATCCATGCCATCAGTCTTGACGTGGACCAGTGTCAGGAGTACGCTGATACCATTCTGCAACGGGTGGCCGATATTAAGAAGCAGTCGTCTTCCGGTGGTAATGGAGGTTCGGGAACTTCGTCAGGCGTCATATCACGCCTCCCCTCCATCGACCTGCCGATCTTCCACGGTGATCTGAACGAGTGGGTCGGCTTTATTAATTTGTTTGACAGCCTGGTCCATGGTCGGTCGGACTTGACTGCCTCTTATAAGATGGCTCAGCTTCGGGGTGCTCTGCGAGGTGAACCTGGTGAATTGGTGGCACACTTGCCTATCACCAACGACAATTATGCGGTCGCACGCCAGATCCTGTTCGACCGATATCAAAATCAACGACGGTTGGTAGACACTCAGCTGGCCAGGCTGTTTGCGATACCCAAACTTTCTCGGGCTTCCGATATCAGAGCCGAGGTACTGAATCCAGTTACGGTGGCCACTAAGGCCTTGGGAAATTTGGGTCTACCAGTAGACCAATGGTCATATCTGCTCTTTTATATTGTTGCAGGTAGGCTGCCGGTTGAGGTGCTCACTCGATTCGAGCAGCAGGACAACGCAGGTAGGGACGAACTGCCTACCCTGGCCAGTCTGCTGGCATTTTTAGAAGCTGAAAGTCGCCGACACGAGATTATGCCACCGCCCTCCTCCTCTGGGTCGCGTGTGCCGGCTTCCGGCCACGCCGTTTCGGTTGCTCAGAGGGGAGGGGGAGGGGATAAGAGGGGGCCGTCTCAACGACTGACTGCGCTCATGGCAGCGGTGGGAGGCCCACGGTGTGCCTTCTGTCATGAGAGCGGGCACGACGTAGCCGCCTGTCCGAGGTTCCGGTCGACGCGCATCAGGGGTAGACGCAACATCGTTGCCCAGCGGCGATGGTGCTTTTTTTGTTTTGGGCCGCACACGGCGAGCGTCTGTCCACAGCCACAATTGTGCCCTAATTGCGACGGTCGGCACCATGCCCTCCTGTGTCTGGAGTCTGCCGCCACTCCGAGGTCATCCGGACGCAGTCATCCAGCCTCGCCGACCGGTGAGCGCACCGGTCGTAGCCCTCGCCAATTCGGCGGCGTGGACCCTCGGAAAGAGGCTTCGCAGGGAGGCGCCGGGTTAGGTACTGCGGGCGGGTGCCAGCGGATGCCCACTAGCTACGCCGGGGTGGCCACGTTGTCCCCCCGTGCGGGGCCGTGTTCGCCTGCTGGATCGAGTAGCCCTGCTCACTCGTCTCCCCCTCCCTCAGAAGTACGGTTTTCCCCTCCGCTGAGGGAGCGGCCGACGCTGGAGCGACTCCCTCCAGTGTTTGGCCCGTTTGGCCATCGTACGCGGAGTGTTGCCCCAGCCCCAGAAGCTGTTTTACTTCCTACCGCTTCTATTCGGGTGTTGGATTCGGATGGCAACACTGTGTTTGCTCGAGCTCTGTTGGATTCCGGCGCACAGGGCAGTATTATGTCGAGCAACCTTGCAGTACGGTTGGGCAATCCTATTTACAAGTCTTCTTATTCTATTCGTGGAGTGGGCAATGTTAGAATTTCGGACGTGATCGGTCACACTTCATTTATTTTCTCATCGAATCCTAACCCTCGCTTACAGTTTAGAGTCTCAGCTTTGGTGATGCCTGAGGTGATAGGTCGTCACCCTCTGGTGAAGGTAAATAGTTACATACGGTCATTGACTTCAGACTTACGGCTAGCGGACCCCAAGTTTGACACACCGGGGCCAGTAGACGTACTACTTGGGGCGGATGTACTAGGGAAGCTGCTCCTGACTGGGAAGCGCGTTCTTCATGCGGAGGGTTTGGTTGCAATGGACACAAAATTAGGGCATGTATTGTTGGGTCCTGCGTTCCGGCCGGTCGCTGCGAGGAAATCGGACAATTTTCTGGTCGGGTCTACGCTTTCGGAAGTAGTCCAACGGTTTTGGGAACTAGAAGAACCTCCTACAGCTTATCGAGTTAACCCGGATGAGGAGGAATGTGAGCTGTTTTACCAGAATAATACGGGTCGTCTCTATTCGGGCCGGTTCTTGTTAAGACTTCCTAAGTCTAGGCGCGACGCGGTGTTCCGTCAGAAATATGTAGAGTTTATGCGGGAGTATGAATCCTTAGGTCACATGTCGGTTTCCAAGGTAGATTGGAGTGCTACGGAGCACTGTTTCCTGCCTCATCATGCGGTTTTGAAAACTCCTAACGGTAAAATTAGGGTCGTGTTTGACGGTTCGGTCACGACCTCCTCCGGCGTCTCGTTAAATCAGTGTCTCCATTCGGGTCCAAAGCTGAACCGGGACATTTCAGACATACTGACGAGCTTCCGCAGGCACCAGATCGTTTTCGTGGCAGACATCCGTATGATGTTCAGGCAAACGGTGATTCATCCGGAGGATAGGCGGTATCAACTCATTCTCTGGCGTGAAAGCTCAGATGAGCCCATAAAAATTTATGAACTGAACACGAACACTTACGGGTTGCGGTCGAGTCCATATTTAGCGGTTCGTTCTTTGCGGGAGTTGGCAGTGAGAGAACGGTTGCACTACCCTCGTGCTGCGGACATTTTGGAACGGGACCTATATGTCGATGACGTATGTACGGGCGCCGACAGTGTCGAAGAGGCCTTATCACGCAAGGATGAACTGATCCGCCTTTTGAGCAGGGGCGGGTACGAGTTGAGAAAATGGCTGTCTAACTGTCCTGAGTTGTTGACTGGTTTGCCAACGGAGTTTCAACAGGACCCCCATCTTTTCGAGAATGTCGATAACCCTAACATGCTGTCGGTCCTAGGGATTCAATATCGTCCGATTCAAGACGAATTTACCTATCGGGTGGAGTTGGAAAATCCAAAGGTATGGTCTAAGCGAACGGTGCTTTCCACAATTGCTCGAACCTTCGATCCAAACGGTTGGATCGCTCCTGTCACTTTTCTCTTAAAGTGCTTCATGCAGCGACTGTGGTCTGCTAACCTGTCATGGGACGAGGGGATTGCTGGTGACCTATTGAAGGATTGGTTGAACTTTTTCTCGTCTCTCCCTGATATTAACCACGTGTCAATACCTCGGAAGCTGGTTCCTGCGGGCAAATACAAGGCGTCTCTCCACGGGTTTTCGGATGCCTCGGAACGCGGATTCGCAGCTGCGGTATACTTGCGAACGGTTTCTTCAACTGGAAGGGTTGATATACGGTTGGTGTTAGCTAAGAGTAAGGTCGCTCCGCTTCGGACACGCATGACGATTCCGAAGCTCGAGTTGTCTGGTGCCGCTCTGTTGACTAGGTTGTTAAACCATACAGCTACTAGTTTACGGTCGGTTATTGATTTAGAAAACACTGTCTATGCTTGGACAGACAGCCAGATAGTGTTGTGCTGGTTGAAGGCTTCGGTTCATACACTTGAAGTTTTCGTGGCCAACCGTGTCTCCCAGATTCAAGGTTCTGAGGTCCCTCTAACATGGAGACACGTGCCTGGTGAGCTCAATCCGGCGGACTGCGCTTCTCGGGGATGCTTAGCATCGGAAATAGTTGATCATCAGTTATGGTGGGGTCCCCAGTGGCTCACCAAGAAAGCTTCTCAGTGGCCTCCCTCTCGCATAGAAGTTCCTCCAGGTCTCGTGGCTGCACCGGCGCAGTTCGAGGATCGAGGAGTGATTCAGGGGTTGGATTTGGATCTGCTCATCAACCGTTATAGCTCCTTGGACAAGCTGGTTAAGGTTACCGCCTGGATTTTGAGGTTTATTCGGAATTGTCGTTTGTCGGTGTCTCAACGCAATATGACACCTGTTGTGTGTCCCATCGAGCGGAAGAAGGCGCTGCTGAAATTAATTGAAGTTGTGCAAGCCACCAGCTTTCATTCAGAATTGAAGGCGCTAAGGACTAAGCGCCCTAAGCTTCGTGGAGCTATCGCTCGTCTTAGCCCCTTTGTAGACGGCCAGGGGCTGATTCGAGTGGGAGGTAGATTAAGTAACTCCAATTTACCTTATTCGGCGCGCCATCCTCTTTTACTTCCCAAGAAGAGCCAGTTAGTTGACCTGTTGGTGATGGATCGCCACATCGCAAATTCGCATTCCGGTTGCAATGCGCTGTTGGCATCCCTCCAGAGGGAATTTTGGATTCTGTCAGGGCGTCGTACGGTTCGGAGCGTCCTTTTCCGGTGTTTACCCTGTTATCGGCTTAAGGCGTCTACGATGCAGCCTCAGATGGGAGATCTGCCTCCAGACAGAGTGAAGAAGATGAGGCCGTTTTCTGGAGTTGGTACGGACTTCGCTGGGCCTTTCATGATCAAGGCTTCACGTTTGCGGAATGTTAGGTTAATTAAGGCTTATCTGTGCGTTTTTGTCTGCCTTTCAACCAAAGCTGTACATCTTGAAGTTGTTGCCGACCTTACAACTGAAGCCTTTATTGCGAGCTTGGATAGATTTGTGTCCCGTCGAGGCTTACCCGAATTGATACGCTCCGATAACGGAACGAATTTTGTAGGTGCGGACCGTTACTTGCGGGATGTAGTAAATTTCTTGAACAATAACCAGGTGGATATTGAAACGGCCCTCTCTCGTCGCGGTATTCGGTGGACCTTCAGTCCTCCAGGATGCCCCCACTGGGGTGGAATTTTCGAGGCAGTAGTAAAATCGGCTAAGACTCACTTGATGCGCGTGATAGGGCAAACCTCGCTCACCTTTGAGGAGTTAACTACGGTATTTTGTAAAATTGAAGCGGTACTTAATTCGCGGCCGTTGTGCCCGCTCAGTTCAGACCCGAATGACTTGGAGTCGTTAACTCCAGGCCATTTCTTGATAGGACAGCCACTCAATGCGTTGCCGGAGTATCCCCTCTCGGATATCAAGCCTGGACGGTTGAGGCGATACCAGATGTTGCAGCAGATGTCCCAAGATTTTTGGAAACGTTGGTCCCTTGAGTATTTACATCTGCTCCAGCAGCGCTTTAAGTGGACGGATAGAACCAGTCCCCCTCACGTCGGTGACCTTGTGTTGGTTAAGGATGCTAACCTGCCGCCACTGCGTTGGCGTAGGGGGCGCATTGTTAATCTGTTTCCCGGTAAGGATGGGACCCCTCGGTCTGCAGAGGTTCTGGTCGGGGACTCGGTTCTCAAACGAGCGGTCACAACGTTGTCCCGGCTGCCAGTTGATTAG
Protein
MSVSYIRGKVTSELRWFASAHALVDRTDRDAALLDRASQVHGRLEVVYDRLSDLVALGSEVEKEVIHAISLDVDQCQEYADTILQRVADIKKQSSSGGNGGSGTSSGVISRLPSIDLPIFHGDLNEWVGFINLFDSLVHGRSDLTASYKMAQLRGALRGEPGELVAHLPITNDNYAVARQILFDRYQNQRRLVDTQLARLFAIPKLSRASDIRAEVLNPVTVATKALGNLGLPVDQWSYLLFYIVAGRLPVEVLTRFEQQDNAGRDELPTLASLLAFLEAESRRHEIMPPPSSSGSRVPASGHAVSVAQRGGGGDKRGPSQRLTALMAAVGGPRCAFCHESGHDVAACPRFRSTRIRGRRNIVAQRRWCFFCFGPHTASVCPQPQLCPNCDGRHHALLCLESAATPRSSGRSHPASPTGERTGRSPRQFGGVDPRKEASQGGAGLGTAGGCQRMPTSYAGVATLSPRAGPCSPAGSSSPAHSSPPPSEVRFSPPLRERPTLERLPPVFGPFGHRTRSVAPAPEAVLLPTASIRVLDSDGNTVFARALLDSGAQGSIMSSNLAVRLGNPIYKSSYSIRGVGNVRISDVIGHTSFIFSSNPNPRLQFRVSALVMPEVIGRHPLVKVNSYIRSLTSDLRLADPKFDTPGPVDVLLGADVLGKLLLTGKRVLHAEGLVAMDTKLGHVLLGPAFRPVAARKSDNFLVGSTLSEVVQRFWELEEPPTAYRVNPDEEECELFYQNNTGRLYSGRFLLRLPKSRRDAVFRQKYVEFMREYESLGHMSVSKVDWSATEHCFLPHHAVLKTPNGKIRVVFDGSVTTSSGVSLNQCLHSGPKLNRDISDILTSFRRHQIVFVADIRMMFRQTVIHPEDRRYQLILWRESSDEPIKIYELNTNTYGLRSSPYLAVRSLRELAVRERLHYPRAADILERDLYVDDVCTGADSVEEALSRKDELIRLLSRGGYELRKWLSNCPELLTGLPTEFQQDPHLFENVDNPNMLSVLGIQYRPIQDEFTYRVELENPKVWSKRTVLSTIARTFDPNGWIAPVTFLLKCFMQRLWSANLSWDEGIAGDLLKDWLNFFSSLPDINHVSIPRKLVPAGKYKASLHGFSDASERGFAAAVYLRTVSSTGRVDIRLVLAKSKVAPLRTRMTIPKLELSGAALLTRLLNHTATSLRSVIDLENTVYAWTDSQIVLCWLKASVHTLEVFVANRVSQIQGSEVPLTWRHVPGELNPADCASRGCLASEIVDHQLWWGPQWLTKKASQWPPSRIEVPPGLVAAPAQFEDRGVIQGLDLDLLINRYSSLDKLVKVTAWILRFIRNCRLSVSQRNMTPVVCPIERKKALLKLIEVVQATSFHSELKALRTKRPKLRGAIARLSPFVDGQGLIRVGGRLSNSNLPYSARHPLLLPKKSQLVDLLVMDRHIANSHSGCNALLASLQREFWILSGRRTVRSVLFRCLPCYRLKASTMQPQMGDLPPDRVKKMRPFSGVGTDFAGPFMIKASRLRNVRLIKAYLCVFVCLSTKAVHLEVVADLTTEAFIASLDRFVSRRGLPELIRSDNGTNFVGADRYLRDVVNFLNNNQVDIETALSRRGIRWTFSPPGCPHWGGIFEAVVKSAKTHLMRVIGQTSLTFEELTTVFCKIEAVLNSRPLCPLSSDPNDLESLTPGHFLIGQPLNALPEYPLSDIKPGRLRRYQMLQQMSQDFWKRWSLEYLHLLQQRFKWTDRTSPPHVGDLVLVKDANLPPLRWRRGRIVNLFPGKDGTPRSAEVLVGDSVLKRAVTTLSRLPVD
Summary
Uniprot
A0A087T9T6
A0A1U8N896
J9JNP4
A0A0J7KBS8
A0A1B6KZV2
A0A1Y1KMX0
+ More
A0A1B6MUZ9 J9JKU0 A0A1Y1LIG5 V5GHM7 A0A0J7KB34 A0A226D417 J9JUK4 A0A0J7KBZ0 A0A2S2NBR8 A0A0J7KCB9 A0A226E226 X1WTX8 A0A0J7KDN6 X1XTX7 X1WZF3 A0A0J7N890 J9JLG6 X1WY95 X1XU26 A0A2S2P1X6 A0A0A9WE49 A0A226CWA5 A0A226DMG9 A0A2S2NDE3 A0A2S2PNC2 A0A0A9VSF7 J9LPA9 A0A2S2PI15 J9L9H5 A0A0J7KCR4 A0A0A9WP39 A0A226EUF5 A0A2S2NNC8 J9JUM9 X1WYQ8 A0A0J7KJY9 J9K351 A0A0J7N536 A0A0J7N5C9 A0A2S2R2J0 A0A0K8SX96 A0A1B6LLT3 A0A1Y1LBZ3 A0A0J7KGI8 A0A1B6MT91 J9JVZ7 A0A0J7K7L1 A0A2S2P963 A0A182HA33 A0A0J7N465 J9LHB7 A0A224XGH3 A0A0J7KCI2 J9K0A1 A0A2S2NPW7 A0A2S2QWS1 A0A226EPI5 A0A2S2NCY5 J9JVD6 A0A1S4EIX5 A0A1Y1L369 J9LIW1 A0A2S2QPS5 A0A1Y1MTT1 X1WZY4 A0A2S2PNZ4 A0A2S2PM34 A0A0A9Z275 A0A2S2PC00 A0A224XG39 J9JUA7 A0A0A9VYL4 A0A226EBC2 A0A023EYK8 A0A3L8DF49 A0A2S2PC01 A0A0A9YU87 X1X5R1 A0A146LBU6 J9JT64 J9KZG0 J9L3C9 A0A2S2PUJ2 X1X8C2 A0A2S2NLW5 X1WYQ7 A0A0A9XKM2 A0A1W7R6I7 A0A2S2NZ32 J9K3V8
A0A1B6MUZ9 J9JKU0 A0A1Y1LIG5 V5GHM7 A0A0J7KB34 A0A226D417 J9JUK4 A0A0J7KBZ0 A0A2S2NBR8 A0A0J7KCB9 A0A226E226 X1WTX8 A0A0J7KDN6 X1XTX7 X1WZF3 A0A0J7N890 J9JLG6 X1WY95 X1XU26 A0A2S2P1X6 A0A0A9WE49 A0A226CWA5 A0A226DMG9 A0A2S2NDE3 A0A2S2PNC2 A0A0A9VSF7 J9LPA9 A0A2S2PI15 J9L9H5 A0A0J7KCR4 A0A0A9WP39 A0A226EUF5 A0A2S2NNC8 J9JUM9 X1WYQ8 A0A0J7KJY9 J9K351 A0A0J7N536 A0A0J7N5C9 A0A2S2R2J0 A0A0K8SX96 A0A1B6LLT3 A0A1Y1LBZ3 A0A0J7KGI8 A0A1B6MT91 J9JVZ7 A0A0J7K7L1 A0A2S2P963 A0A182HA33 A0A0J7N465 J9LHB7 A0A224XGH3 A0A0J7KCI2 J9K0A1 A0A2S2NPW7 A0A2S2QWS1 A0A226EPI5 A0A2S2NCY5 J9JVD6 A0A1S4EIX5 A0A1Y1L369 J9LIW1 A0A2S2QPS5 A0A1Y1MTT1 X1WZY4 A0A2S2PNZ4 A0A2S2PM34 A0A0A9Z275 A0A2S2PC00 A0A224XG39 J9JUA7 A0A0A9VYL4 A0A226EBC2 A0A023EYK8 A0A3L8DF49 A0A2S2PC01 A0A0A9YU87 X1X5R1 A0A146LBU6 J9JT64 J9KZG0 J9L3C9 A0A2S2PUJ2 X1X8C2 A0A2S2NLW5 X1WYQ7 A0A0A9XKM2 A0A1W7R6I7 A0A2S2NZ32 J9K3V8
EMBL
KK114194
KFM61875.1
ABLF02005684
LBMM01009888
KMQ87813.1
GEBQ01022991
+ More
GEBQ01006185 JAT16986.1 JAT33792.1 GEZM01085145 GEZM01085143 JAV60187.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 ABLF02021785 GEZM01054765 JAV73413.1 GALX01004947 JAB63519.1 LBMM01010405 KMQ87487.1 LNIX01000036 OXA39969.1 ABLF02039606 LBMM01009964 KMQ87756.1 GGMR01002024 MBY14643.1 LBMM01009645 KMQ87967.1 LNIX01000007 OXA51795.1 ABLF02054908 LBMM01009202 KMQ88311.1 ABLF02006304 ABLF02036268 ABLF02041614 LBMM01008464 KMQ88855.1 ABLF02030698 ABLF02030705 ABLF02030709 ABLF02030854 ABLF02067347 ABLF02001506 ABLF02008029 ABLF02035899 ABLF02037719 ABLF02046327 ABLF02064627 GGMR01010723 MBY23342.1 GBHO01038805 GBHO01038804 GBHO01038801 JAG04799.1 JAG04800.1 JAG04803.1 LNIX01000069 OXA36894.1 LNIX01000016 OXA46198.1 GGMR01002187 MBY14806.1 GGMR01018294 MBY30913.1 GBHO01044317 GBHO01044315 JAF99286.1 JAF99288.1 ABLF02035780 GGMR01016460 MBY29079.1 ABLF02008636 ABLF02065931 LBMM01009551 KMQ88039.1 GBHO01034065 GBHO01034061 JAG09539.1 JAG09543.1 LNIX01000002 OXA60848.1 GGMR01005823 MBY18442.1 ABLF02004035 ABLF02004036 ABLF02004037 ABLF02004038 ABLF02008018 ABLF02008020 ABLF02012635 ABLF02061171 ABLF02028666 ABLF02066713 LBMM01006490 KMQ90572.1 ABLF02025193 LBMM01009898 KMQ87805.1 LBMM01009746 KMQ87900.1 GGMS01014951 MBY84154.1 GBRD01007977 JAG57844.1 GEBQ01015458 JAT24519.1 GEZM01060254 JAV71154.1 LBMM01007859 KMQ89359.1 GEBQ01000825 JAT39152.1 ABLF02015138 LBMM01012188 KMQ86362.1 GGMR01013370 MBY25989.1 JXUM01030959 KQ560903 KXJ80414.1 LBMM01010417 KMQ87475.1 ABLF02022040 ABLF02056888 GFTR01008866 JAW07560.1 LBMM01009688 KMQ87939.1 ABLF02067195 GGMR01006575 MBY19194.1 GGMS01012379 MBY81582.1 OXA59522.1 GGMR01002421 MBY15040.1 ABLF02008594 GEZM01066272 JAV68024.1 ABLF02003736 ABLF02008292 ABLF02008294 ABLF02046909 ABLF02052378 GGMS01010524 MBY79727.1 GEZM01021505 JAV88969.1 ABLF02036131 ABLF02041527 ABLF02064049 GGMR01018506 MBY31125.1 GGMR01017890 MBY30509.1 GBHO01007689 JAG35915.1 GGMR01014351 MBY26970.1 GFTR01008874 JAW07552.1 ABLF02041650 GBHO01044316 JAF99287.1 LNIX01000005 OXA54454.1 GBBI01004479 JAC14233.1 QOIP01000009 RLU19024.1 GGMR01014306 MBY26925.1 GBHO01008418 GBHO01008416 JAG35186.1 JAG35188.1 ABLF02006784 GDHC01014057 JAQ04572.1 ABLF02023842 ABLF02028949 ABLF02058391 ABLF02005616 ABLF02059449 GGMR01020500 MBY33119.1 ABLF02041762 GGMR01005535 MBY18154.1 ABLF02041670 GBHO01023090 JAG20514.1 GEHC01000869 JAV46776.1 GGMR01009854 MBY22473.1 ABLF02006088 ABLF02006089
GEBQ01006185 JAT16986.1 JAT33792.1 GEZM01085145 GEZM01085143 JAV60187.1 GEBQ01007199 GEBQ01000222 JAT32778.1 JAT39755.1 ABLF02021785 GEZM01054765 JAV73413.1 GALX01004947 JAB63519.1 LBMM01010405 KMQ87487.1 LNIX01000036 OXA39969.1 ABLF02039606 LBMM01009964 KMQ87756.1 GGMR01002024 MBY14643.1 LBMM01009645 KMQ87967.1 LNIX01000007 OXA51795.1 ABLF02054908 LBMM01009202 KMQ88311.1 ABLF02006304 ABLF02036268 ABLF02041614 LBMM01008464 KMQ88855.1 ABLF02030698 ABLF02030705 ABLF02030709 ABLF02030854 ABLF02067347 ABLF02001506 ABLF02008029 ABLF02035899 ABLF02037719 ABLF02046327 ABLF02064627 GGMR01010723 MBY23342.1 GBHO01038805 GBHO01038804 GBHO01038801 JAG04799.1 JAG04800.1 JAG04803.1 LNIX01000069 OXA36894.1 LNIX01000016 OXA46198.1 GGMR01002187 MBY14806.1 GGMR01018294 MBY30913.1 GBHO01044317 GBHO01044315 JAF99286.1 JAF99288.1 ABLF02035780 GGMR01016460 MBY29079.1 ABLF02008636 ABLF02065931 LBMM01009551 KMQ88039.1 GBHO01034065 GBHO01034061 JAG09539.1 JAG09543.1 LNIX01000002 OXA60848.1 GGMR01005823 MBY18442.1 ABLF02004035 ABLF02004036 ABLF02004037 ABLF02004038 ABLF02008018 ABLF02008020 ABLF02012635 ABLF02061171 ABLF02028666 ABLF02066713 LBMM01006490 KMQ90572.1 ABLF02025193 LBMM01009898 KMQ87805.1 LBMM01009746 KMQ87900.1 GGMS01014951 MBY84154.1 GBRD01007977 JAG57844.1 GEBQ01015458 JAT24519.1 GEZM01060254 JAV71154.1 LBMM01007859 KMQ89359.1 GEBQ01000825 JAT39152.1 ABLF02015138 LBMM01012188 KMQ86362.1 GGMR01013370 MBY25989.1 JXUM01030959 KQ560903 KXJ80414.1 LBMM01010417 KMQ87475.1 ABLF02022040 ABLF02056888 GFTR01008866 JAW07560.1 LBMM01009688 KMQ87939.1 ABLF02067195 GGMR01006575 MBY19194.1 GGMS01012379 MBY81582.1 OXA59522.1 GGMR01002421 MBY15040.1 ABLF02008594 GEZM01066272 JAV68024.1 ABLF02003736 ABLF02008292 ABLF02008294 ABLF02046909 ABLF02052378 GGMS01010524 MBY79727.1 GEZM01021505 JAV88969.1 ABLF02036131 ABLF02041527 ABLF02064049 GGMR01018506 MBY31125.1 GGMR01017890 MBY30509.1 GBHO01007689 JAG35915.1 GGMR01014351 MBY26970.1 GFTR01008874 JAW07552.1 ABLF02041650 GBHO01044316 JAF99287.1 LNIX01000005 OXA54454.1 GBBI01004479 JAC14233.1 QOIP01000009 RLU19024.1 GGMR01014306 MBY26925.1 GBHO01008418 GBHO01008416 JAG35186.1 JAG35188.1 ABLF02006784 GDHC01014057 JAQ04572.1 ABLF02023842 ABLF02028949 ABLF02058391 ABLF02005616 ABLF02059449 GGMR01020500 MBY33119.1 ABLF02041762 GGMR01005535 MBY18154.1 ABLF02041670 GBHO01023090 JAG20514.1 GEHC01000869 JAV46776.1 GGMR01009854 MBY22473.1 ABLF02006088 ABLF02006089
Proteomes
Pfam
Interpro
IPR008737
Peptidase_asp_put
+ More
IPR021109 Peptidase_aspartic_dom_sf
IPR040676 DUF5641
IPR000477 RT_dom
IPR005312 DUF1759
IPR041373 RT_RNaseH
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR001969 Aspartic_peptidase_AS
IPR041577 RT_RNaseH_2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR008042 Retrotrans_Pao
IPR001995 Peptidase_A2_cat
IPR001878 Znf_CCHC
IPR015416 Znf_H2C2_histone_UAS-bd
IPR036875 Znf_CCHC_sf
IPR000727 T_SNARE_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR040676 DUF5641
IPR000477 RT_dom
IPR005312 DUF1759
IPR041373 RT_RNaseH
IPR036397 RNaseH_sf
IPR041588 Integrase_H2C2
IPR001969 Aspartic_peptidase_AS
IPR041577 RT_RNaseH_2
IPR001584 Integrase_cat-core
IPR012337 RNaseH-like_sf
IPR008042 Retrotrans_Pao
IPR001995 Peptidase_A2_cat
IPR001878 Znf_CCHC
IPR015416 Znf_H2C2_histone_UAS-bd
IPR036875 Znf_CCHC_sf
IPR000727 T_SNARE_dom
Gene 3D
ProteinModelPortal
A0A087T9T6
A0A1U8N896
J9JNP4
A0A0J7KBS8
A0A1B6KZV2
A0A1Y1KMX0
+ More
A0A1B6MUZ9 J9JKU0 A0A1Y1LIG5 V5GHM7 A0A0J7KB34 A0A226D417 J9JUK4 A0A0J7KBZ0 A0A2S2NBR8 A0A0J7KCB9 A0A226E226 X1WTX8 A0A0J7KDN6 X1XTX7 X1WZF3 A0A0J7N890 J9JLG6 X1WY95 X1XU26 A0A2S2P1X6 A0A0A9WE49 A0A226CWA5 A0A226DMG9 A0A2S2NDE3 A0A2S2PNC2 A0A0A9VSF7 J9LPA9 A0A2S2PI15 J9L9H5 A0A0J7KCR4 A0A0A9WP39 A0A226EUF5 A0A2S2NNC8 J9JUM9 X1WYQ8 A0A0J7KJY9 J9K351 A0A0J7N536 A0A0J7N5C9 A0A2S2R2J0 A0A0K8SX96 A0A1B6LLT3 A0A1Y1LBZ3 A0A0J7KGI8 A0A1B6MT91 J9JVZ7 A0A0J7K7L1 A0A2S2P963 A0A182HA33 A0A0J7N465 J9LHB7 A0A224XGH3 A0A0J7KCI2 J9K0A1 A0A2S2NPW7 A0A2S2QWS1 A0A226EPI5 A0A2S2NCY5 J9JVD6 A0A1S4EIX5 A0A1Y1L369 J9LIW1 A0A2S2QPS5 A0A1Y1MTT1 X1WZY4 A0A2S2PNZ4 A0A2S2PM34 A0A0A9Z275 A0A2S2PC00 A0A224XG39 J9JUA7 A0A0A9VYL4 A0A226EBC2 A0A023EYK8 A0A3L8DF49 A0A2S2PC01 A0A0A9YU87 X1X5R1 A0A146LBU6 J9JT64 J9KZG0 J9L3C9 A0A2S2PUJ2 X1X8C2 A0A2S2NLW5 X1WYQ7 A0A0A9XKM2 A0A1W7R6I7 A0A2S2NZ32 J9K3V8
A0A1B6MUZ9 J9JKU0 A0A1Y1LIG5 V5GHM7 A0A0J7KB34 A0A226D417 J9JUK4 A0A0J7KBZ0 A0A2S2NBR8 A0A0J7KCB9 A0A226E226 X1WTX8 A0A0J7KDN6 X1XTX7 X1WZF3 A0A0J7N890 J9JLG6 X1WY95 X1XU26 A0A2S2P1X6 A0A0A9WE49 A0A226CWA5 A0A226DMG9 A0A2S2NDE3 A0A2S2PNC2 A0A0A9VSF7 J9LPA9 A0A2S2PI15 J9L9H5 A0A0J7KCR4 A0A0A9WP39 A0A226EUF5 A0A2S2NNC8 J9JUM9 X1WYQ8 A0A0J7KJY9 J9K351 A0A0J7N536 A0A0J7N5C9 A0A2S2R2J0 A0A0K8SX96 A0A1B6LLT3 A0A1Y1LBZ3 A0A0J7KGI8 A0A1B6MT91 J9JVZ7 A0A0J7K7L1 A0A2S2P963 A0A182HA33 A0A0J7N465 J9LHB7 A0A224XGH3 A0A0J7KCI2 J9K0A1 A0A2S2NPW7 A0A2S2QWS1 A0A226EPI5 A0A2S2NCY5 J9JVD6 A0A1S4EIX5 A0A1Y1L369 J9LIW1 A0A2S2QPS5 A0A1Y1MTT1 X1WZY4 A0A2S2PNZ4 A0A2S2PM34 A0A0A9Z275 A0A2S2PC00 A0A224XG39 J9JUA7 A0A0A9VYL4 A0A226EBC2 A0A023EYK8 A0A3L8DF49 A0A2S2PC01 A0A0A9YU87 X1X5R1 A0A146LBU6 J9JT64 J9KZG0 J9L3C9 A0A2S2PUJ2 X1X8C2 A0A2S2NLW5 X1WYQ7 A0A0A9XKM2 A0A1W7R6I7 A0A2S2NZ32 J9K3V8
Ontologies
GO
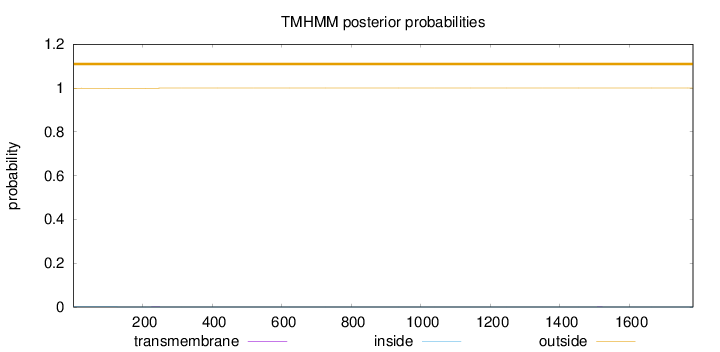
Topology
Length:
1783
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01621
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00061
outside
1 - 1783
Population Genetic Test Statistics
Pi
286.018499
Theta
142.907301
Tajima's D
1.744612
CLR
0.589649
CSRT
0.834508274586271
Interpretation
Uncertain
