Gene
KWMTBOMO12822 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012745
Annotation
PREDICTED:_lipase_1-like_[Bombyx_mori]
Full name
Lipase
Location in the cell
PlasmaMembrane Reliability : 1.613
Sequence
CDS
ATGGTCACGATGATACTCCTGTATCTGAGTAGTACTGGTTATTCACTCACTCCAATTGTGGAAGATCGCTTTTTGGAAAAGCCTGGAGGTGACGTGTTGCAAATTAACGAATTCACGAAAAAATCCGTGTTAAGGGAATTTCCAATACTAGATGAAAACGTGATCAAAACCAATGTACCAGCAGACGCTAAGCTGGACTTCAACGGTCTCGCCGCGAAATACGGACACCCGTGCGAGGAACATTACGTTGAGTCAGAGGACGGTTACGTCACCAAGCTATTTCATTTACCGGGAGACAGGAAACGACCGGTACTCATGATGCACGGCCTCTTCGATTCTGCCGATACTTTTATCATCAGAGGCAACAGATCAATGGCCATATCCCTCGCGAACGCTGGCTACGACGTGTGGCTGGGAAACATGAGAGGAAACAAACATTCCCGGAGACACGTTACTCTGAATCCCGACAAAGATACTGAATTTTGGAACTTCAGTTACCACGAAATGGCTTTGTACGATCTACCTGCGACTATCGACTACATCCTGGGTCACACCGGCCAGACACGGATCTATGGTATCGGTCATTCTCAGGGCAACAATATTTTCATCGTGATGACGGCAATTTTGCCACAATATAACAAAAAGATCAGGGTTTTTATTTCGTTAGCACCGGCAGTCCACATTGAAAATATTAAATTGCCGACGAAAATACTAGTTCTGCTGTCTCCTCTTATAAACTACCTATTCGAAGGGATCGGCAATGAAGAAATTTTTCCCGATGGGACAATTCTTCAGAGGCTACTGCAAAAAGTCTGTCCCTTAGGGTTGCCAGGATATGTTTTATGCGTTCAGTTGATTCTATTTCAACTAGCTGGTTACAATACGGCAGAACTGGAGCAAGAATTCTTCCCGATTGTACTCGGTCATAACCCCAGTGGAACTTCAAGGAAAAACCTTATTCATTTGGCGCAGTCGGGGAAATACAAGCGTTTTGCCTATTACGATGCCGGCGCCGAAGAAAATATCCGATTATATGGAAGAAAGGAACCTCCTTTGTACGATCTCAGCGTGGTCACCATGAAAGTGGCTATTTTTGCTGGAAACAACGACAAATTGGTCGTTCTCAAGGATACTGCAAGACTGCGAGACGAGTTGACAAATGTTGTCGAGTACAGGGTATTGGAACCGAGAAAATGGAATCACTTGGACTTCGTATGGGGGAGGAATGCCAACAAGTTTTTGTTTCCATATATTTTTAAATTGCTGCAGAAATATTAG
Protein
MVTMILLYLSSTGYSLTPIVEDRFLEKPGGDVLQINEFTKKSVLREFPILDENVIKTNVPADAKLDFNGLAAKYGHPCEEHYVESEDGYVTKLFHLPGDRKRPVLMMHGLFDSADTFIIRGNRSMAISLANAGYDVWLGNMRGNKHSRRHVTLNPDKDTEFWNFSYHEMALYDLPATIDYILGHTGQTRIYGIGHSQGNNIFIVMTAILPQYNKKIRVFISLAPAVHIENIKLPTKILVLLSPLINYLFEGIGNEEIFPDGTILQRLLQKVCPLGLPGYVLCVQLILFQLAGYNTAELEQEFFPIVLGHNPSGTSRKNLIHLAQSGKYKRFAYYDAGAEENIRLYGRKEPPLYDLSVVTMKVAIFAGNNDKLVVLKDTARLRDELTNVVEYRVLEPRKWNHLDFVWGRNANKFLFPYIFKLLQKY
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Feature
chain Lipase
Uniprot
H9JT83
H9JYF4
H9JYF5
H9JSY7
A0A194Q1A5
A0A0N1IGF0
+ More
A0A0L7L277 A0A2H1VHX0 A0A1E1WS39 A0A0L7KQV5 A0A2W1BHX8 H9JXK1 A0A212F2S9 A0A194QRY3 H9JSY6 H9JXH3 I4DP30 A0A0L7L5K9 A0A212F0W8 A0A212FGM8 H9JSY5 A0A194Q1X8 H9JSX2 A0A212F2S4 S4PBS0 A0A2A4JBG8 A0A0P4WII3 A0A2A4J9X8 A0A1D2MRG2 A0A151X254 A0A226E1Q5 B0W6G8 A0A182H3N2 W5JF51 A0A2J7QW32 A0A067QLF8 V5I953 A0A1D2MVM7 W5J548 E2B370 Q7QH37 A0A182QF27 A0A1Y9H2R7 B4KKS1 A0A182KX99 A0A1Y9J0S8 A0A1I8JTB8 A0A182FR70 B4KKR3 A0A182SY57 A0A2J7QRG7 A0A151IQS0 A0A067QXT1 A0A195E3Z0 A0A0L7KXY5 B4KKR5 A0A1B6C389 B4KKR4 A0A195FK23 A0A194R2Y3 A0A182LC36 B4HWS6 A0A0M4ENQ4 A0A0N1PFF0 A0A182TUN5 Q9VKT2 A0A084W6W2 Q8T3X7 B4JE82 A0A067R0F9 A0A182UTW8 A0A2J7PUL3 A0A195AVW7 B0W6G4 A0A2A4JCC5 A0A195CME3 A0A2H1WUD0 F5HKY3 A0A2A4IVE2 X1WSL9 A0A158P269 B3N5D0 A0A0L0BZZ8 A0A194QDF4 A0A182R885 Q16F27 B4Q9D3 A0A182JMM3 A0A1W7R7P1 A0A182RCW3 A0A0T6BEX7 A0A182LFG8 Q7QBX7 Q17BM4 A0A182XID0 A0A182HTB5 A0A2J7QRJ0 A0A182TPG6 A0A212FB51 B4GSX2
A0A0L7L277 A0A2H1VHX0 A0A1E1WS39 A0A0L7KQV5 A0A2W1BHX8 H9JXK1 A0A212F2S9 A0A194QRY3 H9JSY6 H9JXH3 I4DP30 A0A0L7L5K9 A0A212F0W8 A0A212FGM8 H9JSY5 A0A194Q1X8 H9JSX2 A0A212F2S4 S4PBS0 A0A2A4JBG8 A0A0P4WII3 A0A2A4J9X8 A0A1D2MRG2 A0A151X254 A0A226E1Q5 B0W6G8 A0A182H3N2 W5JF51 A0A2J7QW32 A0A067QLF8 V5I953 A0A1D2MVM7 W5J548 E2B370 Q7QH37 A0A182QF27 A0A1Y9H2R7 B4KKS1 A0A182KX99 A0A1Y9J0S8 A0A1I8JTB8 A0A182FR70 B4KKR3 A0A182SY57 A0A2J7QRG7 A0A151IQS0 A0A067QXT1 A0A195E3Z0 A0A0L7KXY5 B4KKR5 A0A1B6C389 B4KKR4 A0A195FK23 A0A194R2Y3 A0A182LC36 B4HWS6 A0A0M4ENQ4 A0A0N1PFF0 A0A182TUN5 Q9VKT2 A0A084W6W2 Q8T3X7 B4JE82 A0A067R0F9 A0A182UTW8 A0A2J7PUL3 A0A195AVW7 B0W6G4 A0A2A4JCC5 A0A195CME3 A0A2H1WUD0 F5HKY3 A0A2A4IVE2 X1WSL9 A0A158P269 B3N5D0 A0A0L0BZZ8 A0A194QDF4 A0A182R885 Q16F27 B4Q9D3 A0A182JMM3 A0A1W7R7P1 A0A182RCW3 A0A0T6BEX7 A0A182LFG8 Q7QBX7 Q17BM4 A0A182XID0 A0A182HTB5 A0A2J7QRJ0 A0A182TPG6 A0A212FB51 B4GSX2
Pubmed
19121390
26354079
26227816
28756777
22118469
22651552
+ More
23622113 27289101 26483478 20920257 23761445 24845553 20798317 12364791 14747013 17210077 17994087 20966253 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 21347285 26108605 17510324 22936249
23622113 27289101 26483478 20920257 23761445 24845553 20798317 12364791 14747013 17210077 17994087 20966253 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24438588 21347285 26108605 17510324 22936249
EMBL
BABH01034815
BABH01043411
BABH01038893
BABH01038894
KQ459580
KPI99336.1
+ More
KQ460480 KPJ14361.1 JTDY01008561 JTDY01003454 KOB64514.1 KOB69530.1 ODYU01002476 SOQ40012.1 GDQN01001224 JAT89830.1 JTDY01007194 KOB65389.1 KZ150080 PZC73881.1 BABH01032765 AGBW02010690 OWR48036.1 KQ461196 KPJ06301.1 BABH01038896 BABH01032763 AK403268 BAM19670.1 JTDY01002846 KOB70611.1 AGBW02011015 OWR47367.1 AGBW02008642 OWR52889.1 KPI99333.1 BABH01038949 OWR48034.1 GAIX01002709 JAA89851.1 NWSH01002215 PCG68894.1 GDRN01044880 JAI66895.1 PCG68895.1 LJIJ01000666 ODM95481.1 KQ982580 KYQ54458.1 LNIX01000009 OXA50396.1 DS231848 EDS36642.1 JXUM01107700 KQ565165 KXJ71265.1 ADMH02001587 ETN61958.1 NEVH01009768 PNF32784.1 KK853197 KDR09957.1 GALX01003535 JAB64931.1 LJIJ01000475 ODM97066.1 ADMH02002130 ETN58533.1 GL445305 EFN89852.1 AAAB01008820 EAA05393.4 AXCN02000090 CH933807 EDW12735.1 APCN01002227 EDW12727.1 KRG03395.1 NEVH01011896 PNF31184.1 KQ976755 KYN08590.1 KK853131 KDR10945.1 KQ979657 KYN19900.1 JTDY01004649 KOB67911.1 EDW12729.2 GEDC01029317 GEDC01024313 GEDC01024096 GEDC01020251 JAS07981.1 JAS12985.1 JAS13202.1 JAS17047.1 EDW12728.1 KQ981523 KYN40354.1 KQ460855 KPJ11879.1 CH480818 EDW52471.1 CP012523 ALC38095.1 KQ461072 KPJ09861.1 AE014134 AAF52978.1 AGB92892.1 ATLV01020990 KE525311 KFB45956.1 AY089450 AAL90188.1 CH916368 EDW03602.1 KK852827 KDR15388.1 NEVH01021196 PNF20013.1 KQ976731 KYM76202.1 EDS36638.1 NWSH01002126 PCG69082.1 KQ977622 KYN01254.1 ODYU01011128 SOQ56675.1 AAAB01008879 EGK96944.1 NWSH01005801 PCG63937.1 ABLF02039332 ADTU01007002 CH954177 EDV59009.1 JRES01001173 KNC24794.1 KQ459193 KPJ03000.1 CH478517 EAT32843.1 CM000361 CM002910 EDX04575.1 KMY89585.1 GEHC01000530 JAV47115.1 LJIG01001091 KRT85849.1 AAAB01008859 EAA08216.4 CH477320 EAT43642.1 APCN01000436 PNF31187.1 AGBW02009374 OWR50972.1 CH479189 EDW25481.1
KQ460480 KPJ14361.1 JTDY01008561 JTDY01003454 KOB64514.1 KOB69530.1 ODYU01002476 SOQ40012.1 GDQN01001224 JAT89830.1 JTDY01007194 KOB65389.1 KZ150080 PZC73881.1 BABH01032765 AGBW02010690 OWR48036.1 KQ461196 KPJ06301.1 BABH01038896 BABH01032763 AK403268 BAM19670.1 JTDY01002846 KOB70611.1 AGBW02011015 OWR47367.1 AGBW02008642 OWR52889.1 KPI99333.1 BABH01038949 OWR48034.1 GAIX01002709 JAA89851.1 NWSH01002215 PCG68894.1 GDRN01044880 JAI66895.1 PCG68895.1 LJIJ01000666 ODM95481.1 KQ982580 KYQ54458.1 LNIX01000009 OXA50396.1 DS231848 EDS36642.1 JXUM01107700 KQ565165 KXJ71265.1 ADMH02001587 ETN61958.1 NEVH01009768 PNF32784.1 KK853197 KDR09957.1 GALX01003535 JAB64931.1 LJIJ01000475 ODM97066.1 ADMH02002130 ETN58533.1 GL445305 EFN89852.1 AAAB01008820 EAA05393.4 AXCN02000090 CH933807 EDW12735.1 APCN01002227 EDW12727.1 KRG03395.1 NEVH01011896 PNF31184.1 KQ976755 KYN08590.1 KK853131 KDR10945.1 KQ979657 KYN19900.1 JTDY01004649 KOB67911.1 EDW12729.2 GEDC01029317 GEDC01024313 GEDC01024096 GEDC01020251 JAS07981.1 JAS12985.1 JAS13202.1 JAS17047.1 EDW12728.1 KQ981523 KYN40354.1 KQ460855 KPJ11879.1 CH480818 EDW52471.1 CP012523 ALC38095.1 KQ461072 KPJ09861.1 AE014134 AAF52978.1 AGB92892.1 ATLV01020990 KE525311 KFB45956.1 AY089450 AAL90188.1 CH916368 EDW03602.1 KK852827 KDR15388.1 NEVH01021196 PNF20013.1 KQ976731 KYM76202.1 EDS36638.1 NWSH01002126 PCG69082.1 KQ977622 KYN01254.1 ODYU01011128 SOQ56675.1 AAAB01008879 EGK96944.1 NWSH01005801 PCG63937.1 ABLF02039332 ADTU01007002 CH954177 EDV59009.1 JRES01001173 KNC24794.1 KQ459193 KPJ03000.1 CH478517 EAT32843.1 CM000361 CM002910 EDX04575.1 KMY89585.1 GEHC01000530 JAV47115.1 LJIG01001091 KRT85849.1 AAAB01008859 EAA08216.4 CH477320 EAT43642.1 APCN01000436 PNF31187.1 AGBW02009374 OWR50972.1 CH479189 EDW25481.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000037510
UP000007151
UP000218220
+ More
UP000094527 UP000075809 UP000198287 UP000002320 UP000069940 UP000249989 UP000000673 UP000235965 UP000027135 UP000008237 UP000007062 UP000075886 UP000075884 UP000009192 UP000075882 UP000076407 UP000075840 UP000069272 UP000075901 UP000078542 UP000078492 UP000078541 UP000001292 UP000092553 UP000075902 UP000000803 UP000030765 UP000001070 UP000075903 UP000078540 UP000007819 UP000005205 UP000008711 UP000037069 UP000075900 UP000008820 UP000000304 UP000075880 UP000008744
UP000094527 UP000075809 UP000198287 UP000002320 UP000069940 UP000249989 UP000000673 UP000235965 UP000027135 UP000008237 UP000007062 UP000075886 UP000075884 UP000009192 UP000075882 UP000076407 UP000075840 UP000069272 UP000075901 UP000078542 UP000078492 UP000078541 UP000001292 UP000092553 UP000075902 UP000000803 UP000030765 UP000001070 UP000075903 UP000078540 UP000007819 UP000005205 UP000008711 UP000037069 UP000075900 UP000008820 UP000000304 UP000075880 UP000008744
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JT83
H9JYF4
H9JYF5
H9JSY7
A0A194Q1A5
A0A0N1IGF0
+ More
A0A0L7L277 A0A2H1VHX0 A0A1E1WS39 A0A0L7KQV5 A0A2W1BHX8 H9JXK1 A0A212F2S9 A0A194QRY3 H9JSY6 H9JXH3 I4DP30 A0A0L7L5K9 A0A212F0W8 A0A212FGM8 H9JSY5 A0A194Q1X8 H9JSX2 A0A212F2S4 S4PBS0 A0A2A4JBG8 A0A0P4WII3 A0A2A4J9X8 A0A1D2MRG2 A0A151X254 A0A226E1Q5 B0W6G8 A0A182H3N2 W5JF51 A0A2J7QW32 A0A067QLF8 V5I953 A0A1D2MVM7 W5J548 E2B370 Q7QH37 A0A182QF27 A0A1Y9H2R7 B4KKS1 A0A182KX99 A0A1Y9J0S8 A0A1I8JTB8 A0A182FR70 B4KKR3 A0A182SY57 A0A2J7QRG7 A0A151IQS0 A0A067QXT1 A0A195E3Z0 A0A0L7KXY5 B4KKR5 A0A1B6C389 B4KKR4 A0A195FK23 A0A194R2Y3 A0A182LC36 B4HWS6 A0A0M4ENQ4 A0A0N1PFF0 A0A182TUN5 Q9VKT2 A0A084W6W2 Q8T3X7 B4JE82 A0A067R0F9 A0A182UTW8 A0A2J7PUL3 A0A195AVW7 B0W6G4 A0A2A4JCC5 A0A195CME3 A0A2H1WUD0 F5HKY3 A0A2A4IVE2 X1WSL9 A0A158P269 B3N5D0 A0A0L0BZZ8 A0A194QDF4 A0A182R885 Q16F27 B4Q9D3 A0A182JMM3 A0A1W7R7P1 A0A182RCW3 A0A0T6BEX7 A0A182LFG8 Q7QBX7 Q17BM4 A0A182XID0 A0A182HTB5 A0A2J7QRJ0 A0A182TPG6 A0A212FB51 B4GSX2
A0A0L7L277 A0A2H1VHX0 A0A1E1WS39 A0A0L7KQV5 A0A2W1BHX8 H9JXK1 A0A212F2S9 A0A194QRY3 H9JSY6 H9JXH3 I4DP30 A0A0L7L5K9 A0A212F0W8 A0A212FGM8 H9JSY5 A0A194Q1X8 H9JSX2 A0A212F2S4 S4PBS0 A0A2A4JBG8 A0A0P4WII3 A0A2A4J9X8 A0A1D2MRG2 A0A151X254 A0A226E1Q5 B0W6G8 A0A182H3N2 W5JF51 A0A2J7QW32 A0A067QLF8 V5I953 A0A1D2MVM7 W5J548 E2B370 Q7QH37 A0A182QF27 A0A1Y9H2R7 B4KKS1 A0A182KX99 A0A1Y9J0S8 A0A1I8JTB8 A0A182FR70 B4KKR3 A0A182SY57 A0A2J7QRG7 A0A151IQS0 A0A067QXT1 A0A195E3Z0 A0A0L7KXY5 B4KKR5 A0A1B6C389 B4KKR4 A0A195FK23 A0A194R2Y3 A0A182LC36 B4HWS6 A0A0M4ENQ4 A0A0N1PFF0 A0A182TUN5 Q9VKT2 A0A084W6W2 Q8T3X7 B4JE82 A0A067R0F9 A0A182UTW8 A0A2J7PUL3 A0A195AVW7 B0W6G4 A0A2A4JCC5 A0A195CME3 A0A2H1WUD0 F5HKY3 A0A2A4IVE2 X1WSL9 A0A158P269 B3N5D0 A0A0L0BZZ8 A0A194QDF4 A0A182R885 Q16F27 B4Q9D3 A0A182JMM3 A0A1W7R7P1 A0A182RCW3 A0A0T6BEX7 A0A182LFG8 Q7QBX7 Q17BM4 A0A182XID0 A0A182HTB5 A0A2J7QRJ0 A0A182TPG6 A0A212FB51 B4GSX2
PDB
1K8Q
E-value=2.65353e-58,
Score=571
Ontologies
PATHWAY
GO
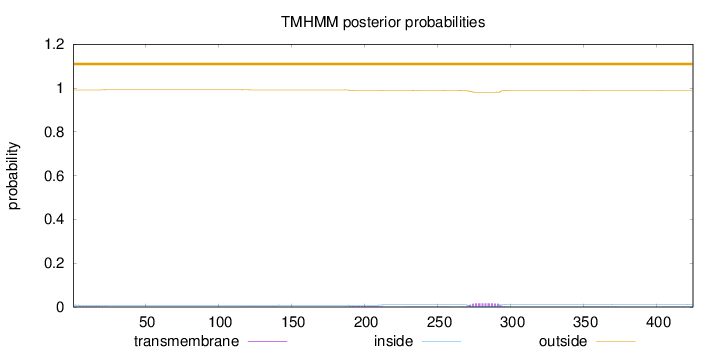
Topology
Length:
425
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.50331
Exp number, first 60 AAs:
0.0253
Total prob of N-in:
0.00922
outside
1 - 425
Population Genetic Test Statistics
Pi
175.839817
Theta
173.494246
Tajima's D
0.081899
CLR
0
CSRT
0.389630518474076
Interpretation
Uncertain
