Gene
KWMTBOMO12820
Annotation
PREDICTED:_neuropeptide_receptor_A35_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.845
Sequence
CDS
ATGACACTTGCTTTCGTAACAAACATTTTCCTTGTGAACCTTTCTGTTGCTGATCTCCTGGTGACCTTGTTCTGCATGCCAGTGCAGATTGCCAAGTCAGTGACCCTGCTGTGGTACTTCGGTGAAGTAATGTGCAAAACCGTCAATTTCCTTCAAGGAGTGGCAGTAGCTTCTAGCGTCTTCACCATAACAGCCATGTCAGTAGATCGGTACTTGGCGATCACTCAGTCCCTTCGGCAACCTTGGATGCCGTCGCGACGTGGCGCGTGCGGACTTCTTGTTTGTCTCTGGCTGATTGCTCTGGCGATCTTCGCGCCGCTGCTTGCTGTGGCGGCAGTTGAACGGGAACGGGTACCATCTTTGTCCAGAACAAAAAATGGATCGATATTTTCCAAATAA
Protein
MTLAFVTNIFLVNLSVADLLVTLFCMPVQIAKSVTLLWYFGEVMCKTVNFLQGVAVASSVFTITAMSVDRYLAITQSLRQPWMPSRRGACGLLVCLWLIALAIFAPLLAVAAVERERVPSLSRTKNGSIFSK
Summary
Miscellaneous
The sequence shown here is derived from an EMBL/GenBank/DDBJ third party annotation (TPA) entry.
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
EMBL
AB330456
BAG68434.1
KZ149907
PZC78269.1
AGBW02007828
OWR54496.1
+ More
RSAL01000176 RVE45033.1 KT031033 ALM88331.1 KQ459324 KPJ01656.1 KQ460416 KPJ14932.1 KPJ01658.1 PZC78270.1 AB330433 BAG68411.1 KT031010 ALM88308.1 NWSH01003879 PCG65728.1 ODYU01001854 SOQ38634.1 KB199650 ESP05214.1 AGBW02014980 OWR40781.1 BK005733 DAA35193.1
RSAL01000176 RVE45033.1 KT031033 ALM88331.1 KQ459324 KPJ01656.1 KQ460416 KPJ14932.1 KPJ01658.1 PZC78270.1 AB330433 BAG68411.1 KT031010 ALM88308.1 NWSH01003879 PCG65728.1 ODYU01001854 SOQ38634.1 KB199650 ESP05214.1 AGBW02014980 OWR40781.1 BK005733 DAA35193.1
Proteomes
Pfam
PF00001 7tm_1
ProteinModelPortal
PDB
5WS3
E-value=1.38434e-14,
Score=187
Ontologies
GO
Topology
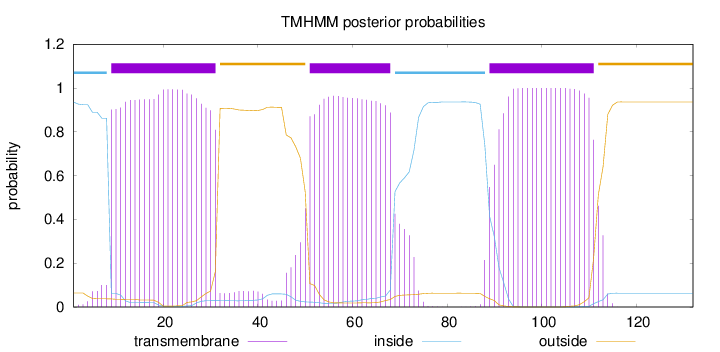
Length:
132
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
65.51667
Exp number, first 60 AAs:
33.60597
Total prob of N-in:
0.93609
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 31
outside
32 - 50
TMhelix
51 - 68
inside
69 - 88
TMhelix
89 - 111
outside
112 - 132
Population Genetic Test Statistics
Pi
209.116543
Theta
162.851802
Tajima's D
0.356136
CLR
13.99613
CSRT
0.469776511174441
Interpretation
Uncertain
